SUPER-FOCUS: a tool for agile functional analysis of shotgun metagenomic data
- PMID: 26454280
- PMCID: PMC4734042
- DOI: 10.1093/bioinformatics/btv584
SUPER-FOCUS: a tool for agile functional analysis of shotgun metagenomic data
Abstract
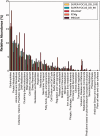
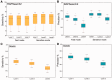
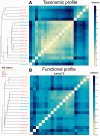
Summary: Analyzing the functional profile of a microbial community from unannotated shotgun sequencing reads is one of the important goals in metagenomics. Functional profiling has valuable applications in biological research because it identifies the abundances of the functional genes of the organisms present in the original sample, answering the question what they can do. Currently, available tools do not scale well with increasing data volumes, which is important because both the number and lengths of the reads produced by sequencing platforms keep increasing. Here, we introduce SUPER-FOCUS, SUbsystems Profile by databasE Reduction using FOCUS, an agile homology-based approach using a reduced reference database to report the subsystems present in metagenomic datasets and profile their abundances. SUPER-FOCUS was tested with over 70 real metagenomes, the results showing that it accurately predicts the subsystems present in the profiled microbial communities, and is up to 1000 times faster than other tools.
Availability and implementation: SUPER-FOCUS was implemented in Python, and its source code and the tool website are freely available at https://edwards.sdsu.edu/SUPERFOCUS.
Contact: redwards@mail.sdsu.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2015. Published by Oxford University Press.
Figures





References
-
- Buchfink B., et al. (2015) Fast and sensitive protein alignment using DIAMOND. Nat. Methods, 12, 59–60. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

