Catalytic strand separation by RECQ1 is required for RPA-mediated response to replication stress
- PMID: 26455304
- PMCID: PMC4631650
- DOI: 10.1016/j.cub.2015.09.026
Catalytic strand separation by RECQ1 is required for RPA-mediated response to replication stress
Abstract
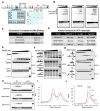
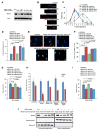
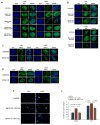
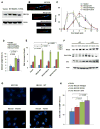
Three (BLM, WRN, and RECQ4) of the five human RecQ helicases are linked to genetic disorders characterized by genomic instability, cancer, and accelerated aging [1]. RECQ1, the first human RecQ helicase discovered [2-4] and the most abundant [5], was recently implicated in breast cancer [6, 7]. RECQ1 is an ATP-dependent DNA-unwinding enzyme (helicase) [8, 9] with roles in replication [10-12] and DNA repair [13-16]. RECQ1 is highly expressed in various tumors and cancer cell lines (for review, see [17]), and its suppression reduces cancer cell proliferation [14], suggesting a target for anti-cancer drugs. RECQ1's assembly state plays a critical role in modulating its helicase, branch migration (BM), or strand annealing [18, 19]. The crystal structure of truncated RECQ1 [20, 21] resembles that of E. coli RecQ [22] with two RecA-like domains, a RecQ-specific zinc-binding domain and a winged-helix domain, the latter implicated in DNA strand separation and oligomer formation. In addition, a conserved aromatic loop (AL) is important for DNA unwinding by bacterial RecQ [23, 24] and truncated RECQ1 helicases [21]. To better understand the roles of RECQ1, two AL mutants (W227A and F231A) in full-length RECQ1 were characterized biochemically and genetically. The RECQ1 mutants were defective in helicase or BM but retained DNA binding, oligomerization, ATPase, and strand annealing. RECQ1-depleted HeLa cells expressing either AL mutant displayed reduced replication tract length, elevated dormant origin firing, and increased double-strand breaks that could be suppressed by exogenously expressed replication protein A (RPA). Thus, RECQ1 governs RPA's availability in order to maintain normal replication dynamics, suppress DNA damage, and preserve genome homeostasis.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
References
-
- Seki M, Yanagisawa J, Kohda T, Sonoyama T, Ui M, Enomoto T. Purification of two DNA-dependent adenosinetriphosphatases having DNA helicase activity from HeLa cells and comparison of the properties of the two enzymes. J Biochem. 1994;115:523–531. - PubMed
-
- Puranam KL, Kennington E, Sait SN, Shows TB, Rochelle JM, Seldin MF, Blackshear PJ. Chromosomal localization of the gene encoding the human DNA helicase RECQL and its mouse homologue. Genomics. 1995;26:595–598. - PubMed
-
- Puranam KL, Blackshear PJ. Cloning and characterization of RECQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J Biol Chem. 1994;269:29838–29845. - PubMed
-
- Kawabe T, Tsuyama N, Kitao S, Nishikawa K, Shimamoto A, Shiratori M, Matsumoto T, Anno K, Sato T, Mitsui Y, et al. Differential regulation of human RecQ family helicases in cell transformation and cell cycle. Oncogene. 2000;19:4764–4772. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

