Intestinal microbiota modulates gluten-induced immunopathology in humanized mice
- PMID: 26456581
- PMCID: PMC4630176
- DOI: 10.1016/j.ajpath.2015.07.018
Intestinal microbiota modulates gluten-induced immunopathology in humanized mice
Abstract
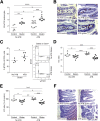
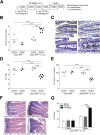
Celiac disease (CD) is an immune-mediated enteropathy triggered by gluten in genetically susceptible individuals. The recent increase in CD incidence suggests that additional environmental factors, such as intestinal microbiota alterations, are involved in its pathogenesis. However, there is no direct evidence of modulation of gluten-induced immunopathology by the microbiota. We investigated whether specific microbiota compositions influence immune responses to gluten in mice expressing the human DQ8 gene, which confers moderate CD genetic susceptibility. Germ-free mice, clean specific-pathogen-free (SPF) mice colonized with a microbiota devoid of opportunistic pathogens and Proteobacteria, and conventional SPF mice that harbor a complex microbiota that includes opportunistic pathogens were used. Clean SPF mice had attenuated responses to gluten compared to germ-free and conventional SPF mice. Germ-free mice developed increased intraepithelial lymphocytes, markers of intraepithelial lymphocyte cytotoxicity, gliadin-specific antibodies, and a proinflammatory gliadin-specific T-cell response. Antibiotic treatment, leading to Proteobacteria expansion, further enhanced gluten-induced immunopathology in conventional SPF mice. Protection against gluten-induced immunopathology in clean SPF mice was reversed after supplementation with a member of the Proteobacteria phylum, an enteroadherent Escherichia coli isolated from a CD patient. The intestinal microbiota can both positively and negatively modulate gluten-induced immunopathology in mice. In subjects with moderate genetic susceptibility, intestinal microbiota changes may be a factor that increases CD risk.
Copyright © 2015 American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures





Comment in
-
Insights into environmental factors impacting celiac disease: microbiota modulation of disease pathogenesis.Am J Pathol. 2015 Nov;185(11):2864-6. doi: 10.1016/j.ajpath.2015.09.001. Epub 2015 Sep 25. Am J Pathol. 2015. PMID: 26404513
References
-
- Dieterich W., Ehnis T., Bauer M., Donner P., Volta U., Riecken E.O., Schuppan D. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat Med. 1997;3:797–801. - PubMed
-
- Shan L., Molberg Ø., Parrot I., Hausch F., Filiz F., Gray G.M., Sollid L.M., Khosla C. Structural basis for gluten intolerance in celiac sprue. Science. 2002;297:2275–2279. - PubMed
-
- Meresse B., Chen Z., Ciszewski C., Tretiakova M., Bhagat G., Krausz T.N., Raulet D.H., Lanier L.L., Groh V., Spies T., Ebert E.C., Green P.H., Jabri B. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity. 2004;21:357–366. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

