Lethal Cardiomyopathy in Mice Lacking Transferrin Receptor in the Heart
- PMID: 26456827
- PMCID: PMC4618069
- DOI: 10.1016/j.celrep.2015.09.023
Lethal Cardiomyopathy in Mice Lacking Transferrin Receptor in the Heart
Abstract
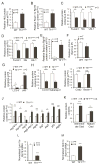
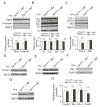
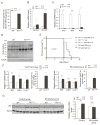
Both iron overload and iron deficiency have been associated with cardiomyopathy and heart failure, but cardiac iron utilization is incompletely understood. We hypothesized that the transferrin receptor (Tfr1) might play a role in cardiac iron uptake and used gene targeting to examine the role of Tfr1 in vivo. Surprisingly, we found that decreased iron, due to inactivation of Tfr1, was associated with severe cardiac consequences. Mice lacking Tfr1 in the heart died in the second week of life and had cardiomegaly, poor cardiac function, failure of mitochondrial respiration, and ineffective mitophagy. The phenotype could only be rescued by aggressive iron therapy, but it was ameliorated by administration of nicotinamide riboside, an NAD precursor. Our findings underscore the importance of both Tfr1 and iron in the heart, and may inform therapy for patients with heart failure.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Transferrin receptor 1-mediated iron uptake plays an essential role in hematopoiesis.Haematologica. 2020 Aug;105(8):2071-2082. doi: 10.3324/haematol.2019.224899. Epub 2019 Oct 10. Haematologica. 2020. PMID: 31601687 Free PMC article.
-
The role of transferrin receptor 1 and 2 in transferrin-bound iron uptake in human hepatoma cells.Am J Physiol Cell Physiol. 2009 Dec;297(6):C1567-75. doi: 10.1152/ajpcell.00649.2008. Epub 2009 Oct 14. Am J Physiol Cell Physiol. 2009. PMID: 19828835
-
Advanced iron-overload cardiomyopathy in a genetic murine model is rescued by resveratrol therapy.Biosci Rep. 2018 Jan 10;38(1):BSR20171302. doi: 10.1042/BSR20171302. Print 2018 Feb 28. Biosci Rep. 2018. PMID: 29208771 Free PMC article.
-
The transferrin receptor: the cellular iron gate.Metallomics. 2017 Oct 18;9(10):1367-1375. doi: 10.1039/c7mt00143f. Metallomics. 2017. PMID: 28671201 Review.
-
[Chrono-drug delivery system based on the circadian rhythm of transferrin receptor].Nihon Rinsho. 2013 Dec;71(12):2200-5. Nihon Rinsho. 2013. PMID: 24437279 Review. Japanese.
Cited by
-
NAD+ homeostasis in health and disease.Nat Metab. 2020 Jan;2(1):9-31. doi: 10.1038/s42255-019-0161-5. Epub 2020 Jan 20. Nat Metab. 2020. PMID: 32694684 Review.
-
Drosophila exercise, an emerging model bridging the fields of exercise and aging in human.Front Cell Dev Biol. 2022 Sep 9;10:966531. doi: 10.3389/fcell.2022.966531. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36158212 Free PMC article. Review.
-
TFRC upregulation promotes ferroptosis in CVB3 infection via nucleus recruitment of Sp1.Cell Death Dis. 2022 Jul 11;13(7):592. doi: 10.1038/s41419-022-05027-w. Cell Death Dis. 2022. PMID: 35821227 Free PMC article.
-
Therapeutic Potential of NAD-Boosting Molecules: The In Vivo Evidence.Cell Metab. 2018 Mar 6;27(3):529-547. doi: 10.1016/j.cmet.2018.02.011. Cell Metab. 2018. PMID: 29514064 Free PMC article. Review.
-
A role for iron deficiency in dopaminergic neurodegeneration.Proc Natl Acad Sci U S A. 2016 Mar 29;113(13):3417-8. doi: 10.1073/pnas.1601976113. Epub 2016 Mar 16. Proc Natl Acad Sci U S A. 2016. PMID: 26984501 Free PMC article. No abstract available.
References
-
- Arany Z, He H, Lin J, Hoyer K, Handschin C, Toka O, Ahmad F, Matsui T, Chin S, Wu PH, et al. Transcriptional coactivator PGC-1 alpha controls the energy state and contractile function of cardiac muscle. Cell Metab. 2005;1:259–271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

