Interindividual methylomic variation across blood, cortex, and cerebellum: implications for epigenetic studies of neurological and neuropsychiatric phenotypes
- PMID: 26457534
- PMCID: PMC4844197
- DOI: 10.1080/15592294.2015.1100786
Interindividual methylomic variation across blood, cortex, and cerebellum: implications for epigenetic studies of neurological and neuropsychiatric phenotypes
Abstract
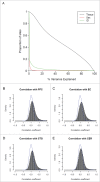
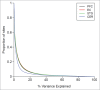
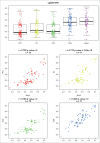
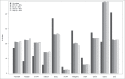
Given the tissue-specific nature of epigenetic processes, the assessment of disease-relevant tissue is an important consideration for epigenome-wide association studies (EWAS). Little is known about whether easily accessible tissues, such as whole blood, can be used to address questions about interindividual epigenomic variation in inaccessible tissues, such as the brain. We quantified DNA methylation in matched DNA samples isolated from whole blood and 4 brain regions (prefrontal cortex, entorhinal cortex, superior temporal gyrus, and cerebellum) from 122 individuals. We explored co-variation between tissues and the extent to which methylomic variation in blood is predictive of interindividual variation identified in the brain. For the majority of DNA methylation sites, interindividual variation in whole blood is not a strong predictor of interindividual variation in the brain, although the relationship with cortical regions is stronger than with the cerebellum. Variation at a subset of probes is strongly correlated across tissues, even in instances when the actual level of DNA methylation is significantly different between them. A substantial proportion of this co-variation, however, is likely to result from genetic influences. Our data suggest that for the majority of the genome, a blood-based EWAS for disorders where brain is presumed to be the primary tissue of interest will give limited information relating to underlying pathological processes. These results do not, however, discount the utility of using a blood-based EWAS to identify biomarkers of disease phenotypes manifest in the brain. We have generated a searchable database for the interpretation of data from blood-based EWAS analyses ( http://epigenetics.essex.ac.uk/bloodbrain/).
Keywords: DNA methylation; Illumina 450K array; blood; brain; cerebellum; cortex; epigenetic epidemiology.
Figures
References
-
- Murphy TM, Mill J. Epigenetics in health and disease: heralding the EWAS era. Lancet 2014; 383:1952-4; PMID:24630775; http://dx.doi.org/10.1016/S0140-6736(14)60269-5 - DOI - PubMed
-
- Heyn H, Carmona FJ, Gomez A, Ferreira HJ, Bell JT, Sayols S, Ward K, Stefansson OA, Moran S, Sandoval J, et al.. DNA methylation profiling in breast cancer discordant identical twins identifies DOK7 as novel epigenetic biomarker. Carcinogenesis 2013; 34:102-8; PMID:23054610; http://dx.doi.org/10.1093/carcin/bgs321 - DOI - PMC - PubMed
-
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al.. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 2009; 41:178-86; PMID:19151715; http://dx.doi.org/10.1038/ng.298 - DOI - PMC - PubMed
-
- Lange CP, Campan M, Hinoue T, Schmitz RF, van der Meulen-de Jong AE, Slingerland H, van der Meulen-de Jong AE, Slingerland H, Kok PJ, van Dijk CM, et al.. Genome-scale discovery of DNA-methylation biomarkers for blood-based detection of colorectal cancer. PLoS One 2012; 7:e50266; PMID:23209692; http://dx.doi.org/10.1371/journal.pone.0050266 - DOI - PMC - PubMed
-
- Liu Y, Aryee MJ, Padyukov L, Fallin MD, Hesselberg E, Runarsson A, Reinius L, Acevedo N, Taub M, Ronninger M, et al.. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol 2013; 31:142-7; PMID:23334450; http://dx.doi.org/10.1038/nbt.2487 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
