Loss of Tifab, a del(5q) MDS gene, alters hematopoiesis through derepression of Toll-like receptor-TRAF6 signaling
- PMID: 26458771
- PMCID: PMC4612089
- DOI: 10.1084/jem.20141898
Loss of Tifab, a del(5q) MDS gene, alters hematopoiesis through derepression of Toll-like receptor-TRAF6 signaling
Abstract
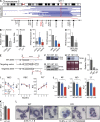
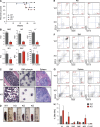
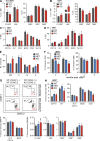
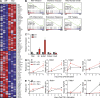
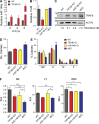
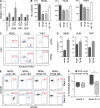
TRAF-interacting protein with forkhead-associated domain B (TIFAB) is a haploinsufficient gene in del(5q) myelodysplastic syndrome (MDS). Deletion of Tifab results in progressive bone marrow (BM) and blood defects, including skewed hematopoietic stem/progenitor cell (HSPC) proportions and altered myeloid differentiation. A subset of mice transplanted with Tifab knockout (KO) HSPCs develop a BM failure with neutrophil dysplasia and cytopenia. In competitive transplants, Tifab KO HSPCs are out-competed by wild-type (WT) cells, suggesting a cell-intrinsic defect. Gene expression analysis of Tifab KO HSPCs identified dysregulation of immune-related signatures, and hypersensitivity to TLR4 stimulation. TIFAB forms a complex with TRAF6, a mediator of immune signaling, and reduces TRAF6 protein stability by a lysosome-dependent mechanism. In contrast, TIFAB loss increases TRAF6 protein and the dynamic range of TLR4 signaling, contributing to ineffective hematopoiesis. Moreover, combined deletion of TIFAB and miR-146a, two genes associated with del(5q) MDS/AML, results in a cooperative increase in TRAF6 expression and hematopoietic dysfunction. Re-expression of TIFAB in del(5q) MDS/AML cells results in attenuated TLR4 signaling and reduced viability. These findings underscore the importance of efficient regulation of innate immune/TRAF6 signaling within HSPCs by TIFAB, and its cooperation with miR-146a as it relates to the pathogenesis of hematopoietic malignancies, such as del(5q) MDS/AML.
© 2015 Varney et al.
Figures



References
-
- Barreyro L., Will B., Bartholdy B., Zhou L., Todorova T.I., Stanley R.F., Ben-Neriah S., Montagna C., Parekh S., Pellagatti A., et al. . 2012. Overexpression of IL-1 receptor accessory protein in stem and progenitor cells and outcome correlation in AML and MDS. Blood. 120:1290–1298. 10.1182/blood-2012-01-404699 - DOI - PMC - PubMed
-
- Boultwood J., Fidler C., Lewis S., Kelly S., Sheridan H., Littlewood T.J., Buckle V.J., and Wainscoat J.S.. 1994. Molecular mapping of uncharacteristically small 5q deletions in two patients with the 5q- syndrome: delineation of the critical region on 5q and identification of a 5q- breakpoint. Genomics. 19:425–432. 10.1006/geno.1994.1090 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

