mTORC2 promotes cell survival through c-Myc-dependent up-regulation of E2F1
- PMID: 26459601
- PMCID: PMC4602034
- DOI: 10.1083/jcb.201411128
mTORC2 promotes cell survival through c-Myc-dependent up-regulation of E2F1
Abstract
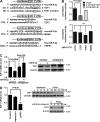
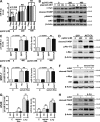
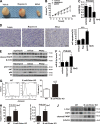
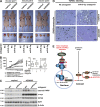
Previous studies have reported that mTORC2 promotes cell survival through phosphorylating AKT and enhancing its activity. We reveal another mechanism by which mTORC2 controls apoptosis. Inactivation of mTORC2 promotes binding of CIP2A to PP2A, leading to reduced PP2A activity toward c-Myc serine 62 and, consequently, enhancement of c-Myc phosphorylation and expression. Increased c-Myc activity induces transcription of pri-miR-9-2/miR-9-3p, in turn inhibiting expression of E2F1, a transcriptional factor critical for cancer cell survival and tumor progression, resulting in enhanced apoptosis. In vivo experiments using B cell-specific mTORC2 (rapamycin-insensitive companion of mTOR) deletion mice and a xenograft tumor model confirmed that inactivation of mTORC2 causes up-regulation of c-Myc and miR-9-3p, down-regulation of E2F1, and consequent reduction in cell survival. Conversely, Antagomir-9-3p reversed mTORC1/2 inhibitor-potentiated E2F1 suppression and resultant apoptosis in xenograft tumors. Our in vitro and in vivo findings collectively demonstrate that mTORC2 promotes cell survival by stimulating E2F1 expression through a c-Myc- and miR-9-3p-dependent mechanism.
© 2015 Zou et al.
Figures





References
-
- Askew D.S., Ashmun R.A., Simmons B.C., and Cleveland J.L.. 1991. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene. 6:1915–1922. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

