Encyclopedia of bacterial gene circuits whose presence or absence correlate with pathogenicity--a large-scale system analysis of decoded bacterial genomes
- PMID: 26459834
- PMCID: PMC4603813
- DOI: 10.1186/s12864-015-1957-7
Encyclopedia of bacterial gene circuits whose presence or absence correlate with pathogenicity--a large-scale system analysis of decoded bacterial genomes
Abstract
Background: Bacterial infections comprise a global health challenge as the incidences of antibiotic resistance increase. Pathogenic potential of bacteria has been shown to be context dependent, varying in response to environment and even within the strains of the same genus.
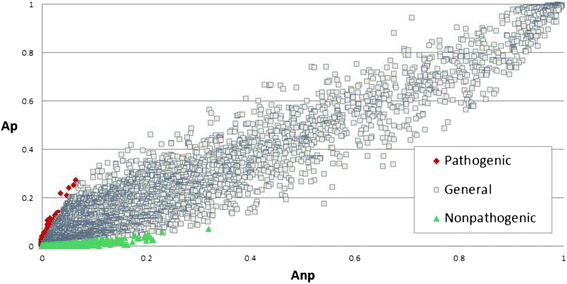
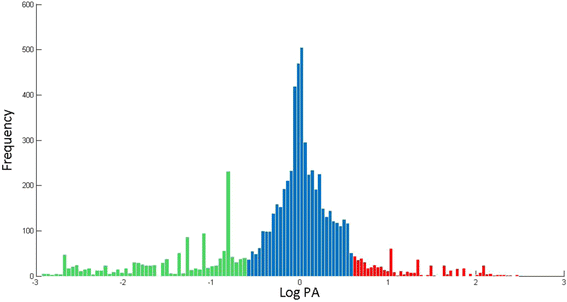
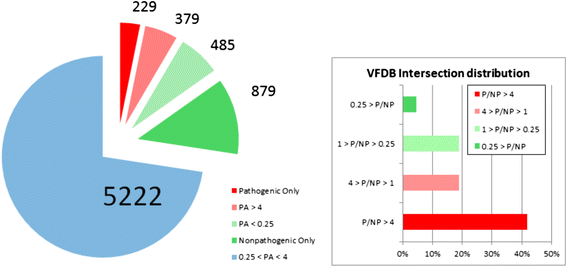
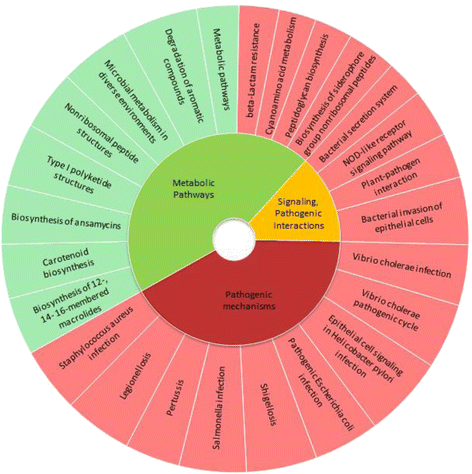
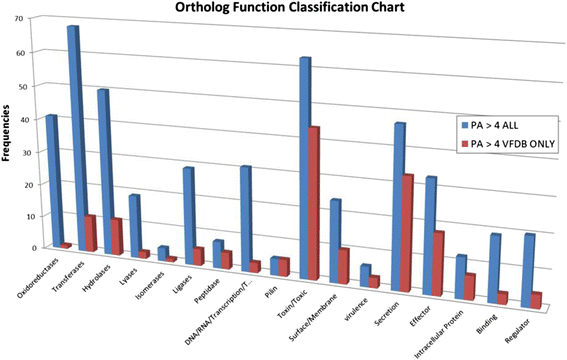
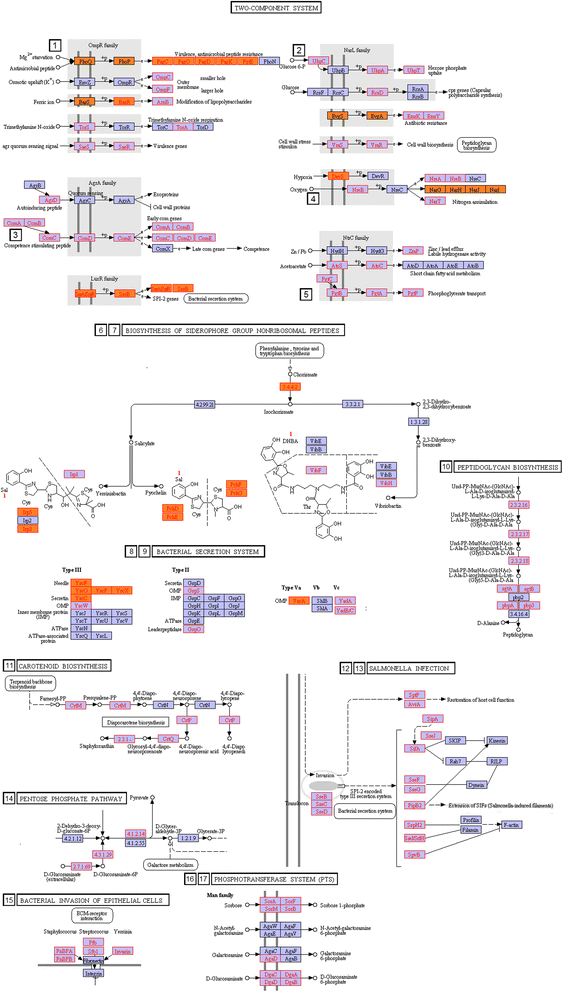
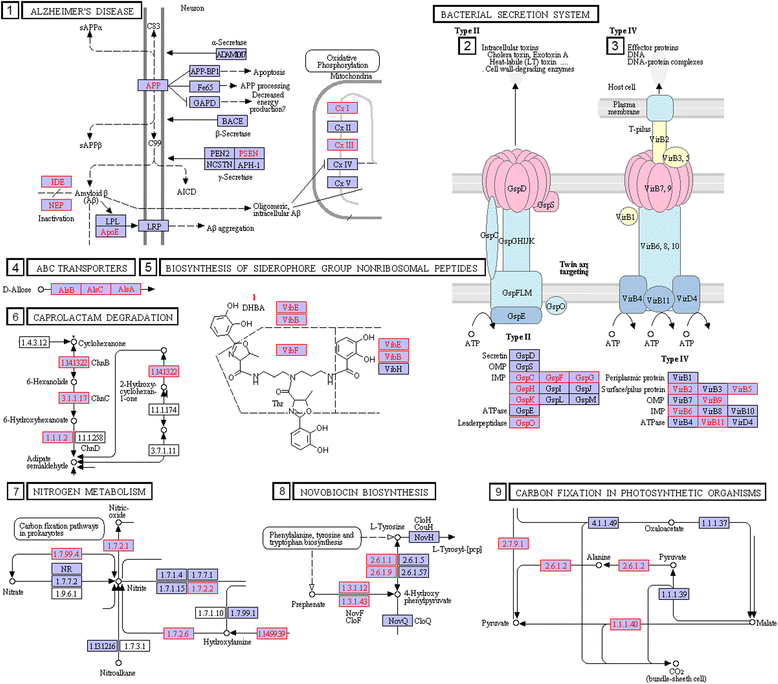
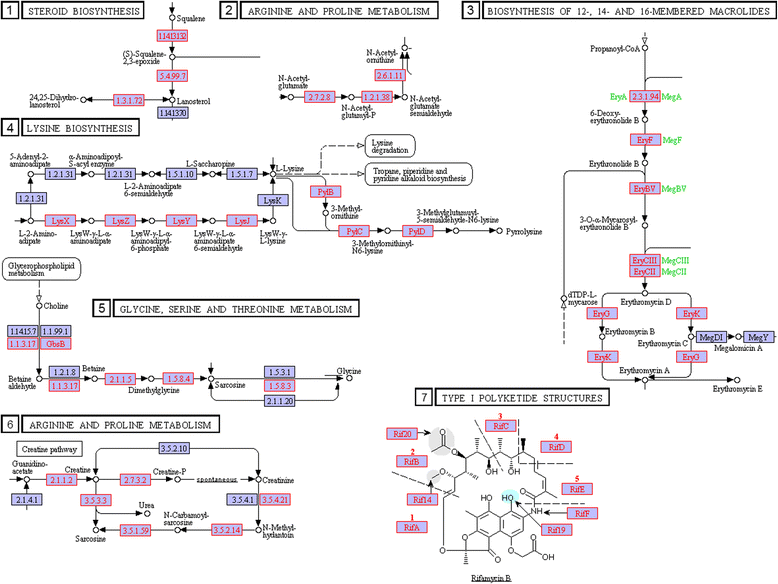
Results: We used the KEGG repository and extensive literature searches to identify among the 2527 bacterial genomes in the literature those implicated as pathogenic to the host, including those which show pathogenicity in a context dependent manner. Using data on the gene contents of these genomes, we identified sets of genes highly abundant in pathogenic but relatively absent in commensal strains and vice versa. In addition, we carried out genome comparison within a genus for the seventeen largest genera in our genome collection. We projected the resultant lists of ortholog genes onto KEGG bacterial pathways to identify clusters and circuits, which can be linked to either pathogenicity or synergy. Gene circuits relatively abundant in nonpathogenic bacteria often mediated biosynthesis of antibiotics. Other synergy-linked circuits reduced drug-induced toxicity. Pathogen-abundant gene circuits included modules in one-carbon folate, two-component system, type-3 secretion system, and peptidoglycan biosynthesis. Antibiotics-resistant bacterial strains possessed genes modulating phagocytosis, vesicle trafficking, cytoskeletal reorganization, and regulation of the inflammatory response. Our study also identified bacterial genera containing a circuit, elements of which were previously linked to Alzheimer's disease.
Conclusions: Present study produces for the first time, a signature, in the form of a robust list of gene circuitry whose presence or absence could potentially define the pathogenicity of a microbiome. Extensive literature search substantiated a bulk majority of the commensal and pathogenic circuitry in our predicted list. Scanning microbiome libraries for these circuitry motifs will provide further insights into the complex and context dependent pathogenicity of bacteria.
Figures
Similar articles
-
Impact of pathogenicity islands in bacterial diagnostics.APMIS. 2004 Nov-Dec;112(11-12):930-6. doi: 10.1111/j.1600-0463.2004.apm11211-1214.x. APMIS. 2004. PMID: 15638844 Review.
-
Using the genome to understand pathogenicity.Methods Mol Biol. 2004;266:261-87. doi: 10.1385/1-59259-763-7:261. Methods Mol Biol. 2004. PMID: 15148423
-
VRprofile: gene-cluster-detection-based profiling of virulence and antibiotic resistance traits encoded within genome sequences of pathogenic bacteria.Brief Bioinform. 2018 Jul 20;19(4):566-574. doi: 10.1093/bib/bbw141. Brief Bioinform. 2018. PMID: 28077405
-
Contribution of genomics to bacterial pathogenesis.Curr Opin Genet Dev. 1999 Dec;9(6):700-3. doi: 10.1016/s0959-437x(99)00021-0. Curr Opin Genet Dev. 1999. PMID: 10607606 Review.
-
A bioinformatic approach to understanding antibiotic resistance in intracellular bacteria through whole genome analysis.Int J Antimicrob Agents. 2008 Sep;32(3):207-20. doi: 10.1016/j.ijantimicag.2008.03.017. Epub 2008 Jul 10. Int J Antimicrob Agents. 2008. PMID: 18619818 Review.
Cited by
-
Structure and Mechanism of d-Glucosaminate-6-phosphate Ammonia-lyase: A Novel Octameric Assembly for a Pyridoxal 5'-Phosphate-Dependent Enzyme, and Unprecedented Stereochemical Inversion in the Elimination Reaction of a d-Amino Acid.Biochemistry. 2021 May 25;60(20):1609-1618. doi: 10.1021/acs.biochem.1c00106. Epub 2021 May 5. Biochemistry. 2021. PMID: 33949189 Free PMC article.
-
LRLSHMDA: Laplacian Regularized Least Squares for Human Microbe-Disease Association prediction.Sci Rep. 2017 Aug 8;7(1):7601. doi: 10.1038/s41598-017-08127-2. Sci Rep. 2017. PMID: 28790448 Free PMC article.
-
Virulence factor activity relationships (VFARs): a bioinformatics perspective.Environ Sci Process Impacts. 2017 Mar 22;19(3):247-260. doi: 10.1039/c6em00689b. Environ Sci Process Impacts. 2017. PMID: 28261716 Free PMC article. Review.
-
Supply of Methionine During Late-Pregnancy Alters Fecal Microbiota and Metabolome in Neonatal Dairy Calves Without Changes in Daily Feed Intake.Front Microbiol. 2019 Sep 19;10:2159. doi: 10.3389/fmicb.2019.02159. eCollection 2019. Front Microbiol. 2019. PMID: 31608024 Free PMC article.
-
Small Molecules That Sabotage Bacterial Virulence.Trends Pharmacol Sci. 2017 Apr;38(4):339-362. doi: 10.1016/j.tips.2017.01.004. Epub 2017 Feb 14. Trends Pharmacol Sci. 2017. PMID: 28209403 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

