Relationship Between Microbiota of the Colonic Mucosa vs Feces and Symptoms, Colonic Transit, and Methane Production in Female Patients With Chronic Constipation
- PMID: 26460205
- PMCID: PMC4727996
- DOI: 10.1053/j.gastro.2015.10.005
Relationship Between Microbiota of the Colonic Mucosa vs Feces and Symptoms, Colonic Transit, and Methane Production in Female Patients With Chronic Constipation
Abstract
Background & aims: In fecal samples from patients with chronic constipation, the microbiota differs from that of healthy subjects. However, the profiles of fecal microbiota only partially replicate those of the mucosal microbiota. It is not clear whether these differences are caused by variations in diet or colonic transit, or are associated with methane production (measured by breath tests). We compared the colonic mucosal and fecal microbiota in patients with chronic constipation and in healthy subjects to investigate the relationships between microbiota and other parameters.
Methods: Sigmoid colonic mucosal and fecal microbiota samples were collected from 25 healthy women (controls) and 25 women with chronic constipation and evaluated by 16S ribosomal RNA gene sequencing (average, 49,186 reads/sample). We assessed associations between microbiota (overall composition and operational taxonomic units) and demographic variables, diet, constipation status, colonic transit, and methane production (measured in breath samples after oral lactulose intake).
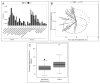
Results: Fourteen patients with chronic constipation had slow colonic transit. The profile of the colonic mucosal microbiota differed between constipated patients and controls (P < .05). The overall composition of the colonic mucosal microbiota was associated with constipation, independent of colonic transit (P < .05), and discriminated between patients with constipation and controls with 94% accuracy. Genera from Bacteroidetes were more abundant in the colonic mucosal microbiota of patients with constipation. The profile of the fecal microbiota was associated with colonic transit before adjusting for constipation, age, body mass index, and diet; genera from Firmicutes (Faecalibacterium, Lactococcus, and Roseburia) correlated with faster colonic transit. Methane production was associated with the composition of the fecal microbiota, but not with constipation or colonic transit.
Conclusions: After adjusting for diet and colonic transit, the profile of the microbiota in the colonic mucosa could discriminate patients with constipation from healthy individuals. The profile of the fecal microbiota was associated with colonic transit and methane production (measured in breath), but not constipation.
Keywords: Irritable Bowel Syndrome; Microbiome.
Copyright © 2016 AGA Institute. Published by Elsevier Inc. All rights reserved.
Figures


Comment in
-
Constipation and the Microbiome: Lumen Versus Mucosa!Gastroenterology. 2016 Feb;150(2):300-3. doi: 10.1053/j.gastro.2015.12.023. Epub 2015 Dec 19. Gastroenterology. 2016. PMID: 26710987 No abstract available.
References
-
- Malinen E, Rinttila T, Kajander K, et al. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100:373–382. - PubMed
-
- Kassinen A, Krogius-Kurikka L, Makivuokko H, et al. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 2007;133:24–33. - PubMed
-
- Tana C, Umesaki Y, Imaoka A, et al. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol Motil. 2010;22:512–519. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

