Current and future prospects for CRISPR-based tools in bacteria
- PMID: 26460902
- PMCID: PMC4816669
- DOI: 10.1002/bit.25851
Current and future prospects for CRISPR-based tools in bacteria
Abstract
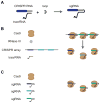
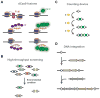
CRISPR-Cas systems have rapidly transitioned from intriguing prokaryotic defense systems to powerful and versatile biomolecular tools. This article reviews how these systems have been translated into technologies to manipulate bacterial genetics, physiology, and communities. Recent applications in bacteria have centered on multiplexed genome editing, programmable gene regulation, and sequence-specific antimicrobials, while future applications can build on advances in eukaryotes, the rich natural diversity of CRISPR-Cas systems, and the untapped potential of CRISPR-based DNA acquisition. Overall, these systems have formed the basis of an ever-expanding genetic toolbox and hold tremendous potential for our future understanding and engineering of the bacterial world.
Keywords: Cas9; antimicrobials; genetic circuits; genetic control; genome engineering; undomesticated microbes.
© 2015 Wiley Periodicals, Inc.
Figures




References
-
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

