Identification and Genome Characterization of the First Sicinivirus Isolate from Chickens in Mainland China by Using Viral Metagenomics
- PMID: 26461027
- PMCID: PMC4603672
- DOI: 10.1371/journal.pone.0139668
Identification and Genome Characterization of the First Sicinivirus Isolate from Chickens in Mainland China by Using Viral Metagenomics
Abstract
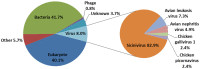
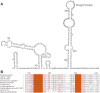
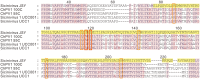
Unlike traditional virus isolation and sequencing approaches, sequence-independent amplification based viral metagenomics technique allows one to discover unexpected or novel viruses efficiently while bypassing culturing step. Here we report the discovery of the first Sicinivirus isolate (designated as strain JSY) of picornaviruses from commercial layer chickens in mainland China by using a viral metagenomics technique. This Sicinivirus isolate, which contains a whole genome of 9,797 nucleotides (nt) excluding the poly(A) tail, possesses one of the largest picornavirus genome so far reported, but only shares 88.83% and 82.78% of amino acid sequence identity to that of ChPV1 100C (KF979332) and Sicinivirus 1 strain UCC001 (NC_023861), respectively. The complete 939 nt 5'UTR of the isolate strain contains at least twelve stem-loop domains (A-L), representing the highest set of loops reported within Sicinivirus genus. The conserved 'barbell-like' structure was also present in the 272 nt 3'UTR of the isolate as that in the 3' UTR of Sicinivirus 1 strain UCC001. The 8,586 nt large open reading frame encodes a 2,862 amino acids polyprotein precursor. Moreover, Sicinivirus infection might be widely present in commercial chicken farms in Yancheng region of the Jiangsu Province as evidenced by all the tested stool samples from three different farms being positive (17/17) for Sicinivirus detection. This is the first report on identification of Sicinivirus in commercial layer chickens with a severe clinical disease in mainland China, however, further studies are needed to evaluate the pathogenic potential of this picornavirus in chickens.
Conflict of interest statement
Figures

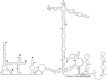

Similar articles
-
Identification and genetic characterization of a novel picornavirus from chickens.J Gen Virol. 2014 May;95(Pt 5):1094-1103. doi: 10.1099/vir.0.061085-0. Epub 2014 Feb 4. J Gen Virol. 2014. PMID: 24496829
-
Identification of a novel bovine enterovirus possessing highly divergent amino acid sequences in capsid protein.BMC Microbiol. 2017 Jan 17;17(1):18. doi: 10.1186/s12866-016-0923-0. BMC Microbiol. 2017. PMID: 28095784 Free PMC article.
-
Whole genome analysis of a novel picornavirus related to the Enterovirus/Sapelovirus supergroup from porcine feces in Japan.Virus Res. 2018 Sep 15;257:68-73. doi: 10.1016/j.virusres.2018.09.003. Epub 2018 Sep 15. Virus Res. 2018. PMID: 30227146
-
[Structure and function of 3'- untranslated region in picornavirus].Bing Du Xue Bao. 2014 Jul;30(4):463-9. Bing Du Xue Bao. 2014. PMID: 25272604 Review. Chinese.
-
Functional analysis of structural motifs in dicistroviruses.Virus Res. 2009 Feb;139(2):137-47. doi: 10.1016/j.virusres.2008.06.006. Epub 2008 Jul 25. Virus Res. 2009. PMID: 18621089 Review.
Cited by
-
Novel Virus Identification through Metagenomics: A Systematic Review.Life (Basel). 2022 Dec 7;12(12):2048. doi: 10.3390/life12122048. Life (Basel). 2022. PMID: 36556413 Free PMC article. Review.
-
Metagenomic sequencing determines complete infectious bronchitis virus (avian Gammacoronavirus) vaccine strain genomes and associated viromes in chicken clinical samples.Virus Genes. 2021 Dec;57(6):529-540. doi: 10.1007/s11262-021-01872-7. Epub 2021 Oct 9. Virus Genes. 2021. PMID: 34626348 Free PMC article.
-
Near-Complete Genome Sequences of Five Siciniviruses from North America.Microbiol Resour Announc. 2021 May 13;10(19):e00364-21. doi: 10.1128/MRA.00364-21. Microbiol Resour Announc. 2021. PMID: 33986098 Free PMC article.
-
A metagenomic comparison of endemic viruses from broiler chickens with runting-stunting syndrome and from normal birds.Avian Pathol. 2016 Dec;45(6):616-629. doi: 10.1080/03079457.2016.1193123. Epub 2016 Oct 4. Avian Pathol. 2016. PMID: 27215546 Free PMC article.
-
A decade of RNA virus metagenomics is (not) enough.Virus Res. 2018 Jan 15;244:218-229. doi: 10.1016/j.virusres.2017.10.014. Epub 2017 Oct 18. Virus Res. 2018. PMID: 29055712 Free PMC article. Review.
References
-
- Boros A, Nemes C, Pankovics P, Kapusinszky B, Delwart E, Reuter G (2013) Genetic characterization of a novel picornavirus in turkeys (Meleagris gallopavo) distinct from turkey galliviruses and megriviruses and distantly related to the members of the genus Avihepatovirus. J Gen Virol 94(Pt 7):1496–1509. 10.1099/vir.0.051797-0 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

