Targeting Human Cancer by a Glycosaminoglycan Binding Malaria Protein
- PMID: 26461094
- PMCID: PMC4790448
- DOI: 10.1016/j.ccell.2015.09.003
Targeting Human Cancer by a Glycosaminoglycan Binding Malaria Protein
Abstract
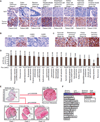
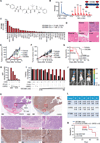
Plasmodium falciparum engineer infected erythrocytes to present the malarial protein, VAR2CSA, which binds a distinct type chondroitin sulfate (CS) exclusively expressed in the placenta. Here, we show that the same CS modification is present on a high proportion of malignant cells and that it can be specifically targeted by recombinant VAR2CSA (rVAR2). In tumors, placental-like CS chains are linked to a limited repertoire of cancer-associated proteoglycans including CD44 and CSPG4. The rVAR2 protein localizes to tumors in vivo and rVAR2 fused to diphtheria toxin or conjugated to hemiasterlin compounds strongly inhibits in vivo tumor cell growth and metastasis. Our data demonstrate how an evolutionarily refined parasite-derived protein can be exploited to target a common, but complex, malignancy-associated glycosaminoglycan modification.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures





References
-
- Alkhalil A, Achur RN, Valiyaveettil M, Ockenhouse CF, Gowda DC. Structural requirements for the adherence of Plasmodium falciparum-infected erythrocytes to chondroitin sulfate proteoglycans of human placenta. J. Biol. Chem. 2000;275:40357–40364. - PubMed
-
- Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, Taraschi TF, Howard RJ. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. - PubMed
-
- Beeson JG, Andrews KT, Boyle M, Duffy MF, Choong EK, Byrne TJ, Chesson JM, Lawson AM, Chai W. Structural basis for binding of Plasmodium falciparum erythrocyte membrane protein 1 to chondroitin sulfate and placental tissue and the influence of protein polymorphisms on binding specificity. J. Biol. Chem. 2007;282:22426–22436. - PubMed
-
- Casucci M, Nicolis di Robilant B, Falcone L, Camisa B, Norelli M, Genovese P, Gentner B, Gullotta F, Ponzoni M, Bernardi M, et al. CD44v6-targeted T cells mediate potent antitumor effects against acute myeloid leukemia and multiple myeloma. Blood. 2013;122:3461–3472. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

