The first two cell-fate decisions of preimplantation mouse embryo development are not functionally independent
- PMID: 26461180
- PMCID: PMC4602213
- DOI: 10.1038/srep15034
The first two cell-fate decisions of preimplantation mouse embryo development are not functionally independent
Abstract
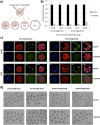
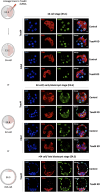
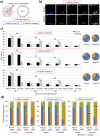
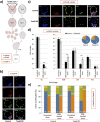
During mouse preimplantation embryo development, three distinct cell lineages are formed, represented by the differentiating trophectoderm (TE), primitive endoderm (PrE) and the pluripotent epiblast (EPI). Classically, lineage derivation has been presented as a two-step process whereby outer TE cells are first segregated from inner-cell mass (ICM), followed by ICM refinement into either the PrE or EPI. As ICM founders can be produced following the fourth or fifth cleavage divisions, their potential to equally contribute to EPI and PrE is contested. Thus, modelling the early sequestration of ICM founders from TE-differentiation after the fourth cleavage division, we examined ICM lineage contribution of varying sized cell clones unable to initiate TE-differentiation. Such TE-inhibited ICM cells do not equally contribute to EPI and PrE and are significantly biased to form EPI. This bias is not caused by enhanced expression of the EPI marker Nanog, nor correlated with reduced apical polarity but associated with reduced expression of PrE-related gene transcripts (Dab2 and Lrp2) and down-regulation of plasma membrane associated Fgfr2. Our results favour a unifying model were the three cell lineages are guided in an integrated, yet flexible, fate decision centred on relative exposure of founder cells to TE-differentiative cues.
Figures

Similar articles
-
Lineage segregation in human pre-implantation embryos is specified by YAP1 and TEAD1.Hum Reprod. 2023 Aug 1;38(8):1484-1498. doi: 10.1093/humrep/dead107. Hum Reprod. 2023. PMID: 37295962
-
Stochastic patterning in the mouse pre-implantation embryo.Development. 2007 Dec;134(23):4219-31. doi: 10.1242/dev.003798. Epub 2007 Oct 31. Development. 2007. PMID: 17978007
-
[Epiblast and primitive endoderm cell specification during mouse preimplantation development: a combination between biology and mathematical modeling].Med Sci (Paris). 2016 Feb;32(2):192-7. doi: 10.1051/medsci/20163202013. Epub 2016 Mar 2. Med Sci (Paris). 2016. PMID: 26936177 Review. French.
-
Formation of distinct cell types in the mouse blastocyst.Results Probl Cell Differ. 2012;55:203-17. doi: 10.1007/978-3-642-30406-4_11. Results Probl Cell Differ. 2012. PMID: 22918808 Review.
-
Allocation of inner cells to epiblast vs primitive endoderm in the mouse embryo is biased but not determined by the round of asymmetric divisions (8→16- and 16→32-cells).Dev Biol. 2014 Jan 1;385(1):136-48. doi: 10.1016/j.ydbio.2013.09.008. Epub 2013 Sep 13. Dev Biol. 2014. PMID: 24041854
Cited by
-
HIPPO signaling resolves embryonic cell fate conflicts during establishment of pluripotency in vivo.Elife. 2018 Dec 11;7:e42298. doi: 10.7554/eLife.42298. Elife. 2018. PMID: 30526858 Free PMC article.
-
p38 (Mapk14/11) occupies a regulatory node governing entry into primitive endoderm differentiation during preimplantation mouse embryo development.Open Biol. 2016 Sep;6(9):160190. doi: 10.1098/rsob.160190. Open Biol. 2016. PMID: 27605380 Free PMC article.
-
Mouse ICM Organoids Reveal Three-Dimensional Cell Fate Clustering.Biophys J. 2019 Jan 8;116(1):127-141. doi: 10.1016/j.bpj.2018.11.011. Epub 2018 Nov 16. Biophys J. 2019. PMID: 30514631 Free PMC article.
-
Trophectoderm formation: regulation of morphogenesis and gene expressions by RHO, ROCK, cell polarity, and HIPPO signaling.Reproduction. 2022 Aug 22;164(4):R75-R86. doi: 10.1530/REP-21-0478. Print 2022 Oct 1. Reproduction. 2022. PMID: 35900353 Free PMC article. Review.
-
The transition from local to global patterns governs the differentiation of mouse blastocysts.PLoS One. 2020 May 15;15(5):e0233030. doi: 10.1371/journal.pone.0233030. eCollection 2020. PLoS One. 2020. PMID: 32413083 Free PMC article.
References
-
- Zernicka-Goetz M., Morris S. A. & Bruce A. W. Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat Rev Genet 10, 467–477 (2009). - PubMed
-
- Johnson M. H. & Ziomek C. A. The foundation of two distinct cell lineages within the mouse morula. Cell 24, 71–80 (1981). - PubMed
-
- Sutherland A. E., Speed T. P. & Calarco P. G. Inner cell allocation in the mouse morula: the role of oriented division during fourth cleavage. Dev Biol 137, 13–25 (1990). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

