Quantitative phosphoproteomics reveals new roles for the protein phosphatase PP6 in mitotic cells
- PMID: 26462736
- PMCID: PMC5244978
- DOI: 10.1126/scisignal.aab3138
Quantitative phosphoproteomics reveals new roles for the protein phosphatase PP6 in mitotic cells
Abstract
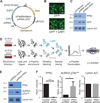
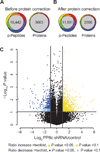
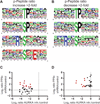
Protein phosphorylation is an important regulatory mechanism controlling mitotic progression. Protein phosphatase 6 (PP6) is an essential enzyme with conserved roles in chromosome segregation and spindle assembly from yeast to humans. We applied a baculovirus-mediated gene silencing approach to deplete HeLa cells of the catalytic subunit of PP6 (PP6c) and analyzed changes in the phosphoproteome and proteome in mitotic cells by quantitative mass spectrometry-based proteomics. We identified 408 phosphopeptides on 272 proteins that increased and 298 phosphopeptides on 220 proteins that decreased in phosphorylation upon PP6c depletion in mitotic cells. Motif analysis of the phosphorylated sites combined with bioinformatics pathway analysis revealed previously unknown PP6c-dependent regulatory pathways. Biochemical assays demonstrated that PP6c opposed casein kinase 2-dependent phosphorylation of the condensin I subunit NCAP-G, and cellular analysis showed that depletion of PP6c resulted in defects in chromosome condensation and segregation in anaphase, consistent with dysregulation of condensin I function in the absence of PP6 activity.
Copyright © 2015, American Association for the Advancement of Science.
Figures


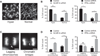

References
-
- Barr FA, Elliott PR, Gruneberg U. Protein phosphatases and the regulation of mitosis. J. Cell Sci. 2011;124:2323–2334. - PubMed
-
- Wurzenberger C, Gerlich DW. Phosphatases: Providing safe passage through mitotic exit. Nat. Rev. Mol. Cell Biol. 2011;12:469–482. - PubMed
-
- Prickett TD, Brautigan DL. The α4 regulatory subunit exerts opposing allosteric effects on protein phosphatases PP6 and PP2A. J. Biol. Chem. 2006;281:30503–30511. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

