2μ plasmid in Saccharomyces species and in Saccharomyces cerevisiae
- PMID: 26463005
- PMCID: PMC4809986
- DOI: 10.1093/femsyr/fov090
2μ plasmid in Saccharomyces species and in Saccharomyces cerevisiae
Abstract
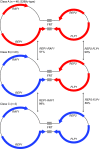
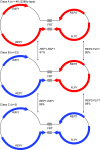
We determined that extrachromosomal 2μ plasmid was present in 67 of the Saccharomyces cerevisiae 100-genome strains; in addition to variation in the size and copy number of 2μ, we identified three distinct classes of 2μ. We identified 2μ presence/absence and class associations with populations, clinical origin and nuclear genotypes. We also screened genome sequences of S. paradoxus, S. kudriavzevii, S. uvarum, S. eubayanus, S. mikatae, S. arboricolus and S. bayanus strains for both integrated and extrachromosomal 2μ. Similar to S. cerevisiae, we found no integrated 2μ sequences in any S. paradoxus strains. However, we identified part of 2μ integrated into the genomes of some S. uvarum, S. kudriavzevii, S. mikatae and S. bayanus strains, which were distinct from each other and from all extrachromosomal 2μ. We identified extrachromosomal 2μ in one S. paradoxus, one S. eubayanus, two S. bayanus and 13 S. uvarum strains. The extrachromosomal 2μ in S. paradoxus, S. eubayanus and S. cerevisiae were distinct from each other. In contrast, the extrachromosomal 2μ in S. bayanus and S. uvarum strains were identical with each other and with one of the three classes of S. cerevisiae 2μ, consistent with interspecific transfer.
Keywords: 2-micron plasmid; 2μ plasmid; Saccharomyces cerevisiae; Saccharomyces genus; population genomics; selfish genetic elements.
© FEMS 2015. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
Similar articles
-
Truncation of Gal4p explains the inactivation of the GAL/MEL regulon in both Saccharomyces bayanus and some Saccharomyces cerevisiae wine strains.FEMS Yeast Res. 2016 Sep;16(6):fow070. doi: 10.1093/femsyr/fow070. Epub 2016 Sep 1. FEMS Yeast Res. 2016. PMID: 27589939
-
Molecular genetic study of introgression between Saccharomyces bayanus and S. cerevisiae.Yeast. 2005 Oct 30;22(14):1099-115. doi: 10.1002/yea.1298. Yeast. 2005. PMID: 16240458
-
New family of pectinase genes PGU1b-PGU3b of the pectinolytic yeast Saccharomyces bayanus var. uvarum.Dokl Biochem Biophys. 2016 Mar;467(1):89-91. doi: 10.1134/S1607672916020034. Epub 2016 May 20. Dokl Biochem Biophys. 2016. PMID: 27193705
-
The ecology and evolution of non-domesticated Saccharomyces species.Yeast. 2014 Dec;31(12):449-62. doi: 10.1002/yea.3040. Epub 2014 Oct 23. Yeast. 2014. PMID: 25242436 Free PMC article. Review.
-
Interspecies hybridization and recombination in Saccharomyces wine yeasts.FEMS Yeast Res. 2008 Nov;8(7):996-1007. doi: 10.1111/j.1567-1364.2008.00369.x. Epub 2008 Mar 18. FEMS Yeast Res. 2008. PMID: 18355270 Review.
Cited by
-
Into the wild: new yeast genomes from natural environments and new tools for their analysis.FEMS Yeast Res. 2020 Mar 1;20(2):foaa008. doi: 10.1093/femsyr/foaa008. FEMS Yeast Res. 2020. PMID: 32009143 Free PMC article. Review.
-
A natural variant of the essential host gene MMS21 restricts the parasitic 2-micron plasmid in Saccharomyces cerevisiae.Elife. 2020 Oct 16;9:e62337. doi: 10.7554/eLife.62337. Elife. 2020. PMID: 33063663 Free PMC article.
-
The yeast 2-μm plasmid Raf protein contributes to plasmid inheritance by stabilizing the Rep1 and Rep2 partitioning proteins.Nucleic Acids Res. 2017 Oct 13;45(18):10518-10533. doi: 10.1093/nar/gkx703. Nucleic Acids Res. 2017. PMID: 29048592 Free PMC article.
-
Truth in wine yeast.Microb Biotechnol. 2022 May;15(5):1339-1356. doi: 10.1111/1751-7915.13848. Epub 2021 Jun 26. Microb Biotechnol. 2022. PMID: 34173338 Free PMC article. Review.
-
Genome Diversity and Evolution in the Budding Yeasts (Saccharomycotina).Genetics. 2017 Jun;206(2):717-750. doi: 10.1534/genetics.116.199216. Genetics. 2017. PMID: 28592505 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

