The no-SCAR (Scarless Cas9 Assisted Recombineering) system for genome editing in Escherichia coli
- PMID: 26463009
- PMCID: PMC4604488
- DOI: 10.1038/srep15096
The no-SCAR (Scarless Cas9 Assisted Recombineering) system for genome editing in Escherichia coli
Abstract
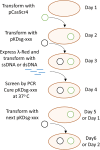
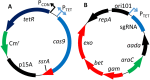
Genome engineering methods in E. coli allow for easy to perform manipulations of the chromosome in vivo with the assistance of the λ-Red recombinase system. These methods generally rely on the insertion of an antibiotic resistance cassette followed by removal of the same cassette, resulting in a two-step procedure for genomic manipulations. Here we describe a method and plasmid system that can edit the genome of E. coli without chromosomal markers. This system, known as Scarless Cas9 Assisted Recombineering (no-SCAR), uses λ-Red to facilitate genomic integration of donor DNA and double stranded DNA cleavage by Cas9 to counterselect against wild-type cells. We show that point mutations, gene deletions, and short sequence insertions were efficiently performed in several genomic loci in a single-step with regards to the chromosome and did not leave behind scar sites. The single-guide RNA encoding plasmid can be easily cured due to its temperature sensitive origin of replication, allowing for iterative chromosomal manipulations of the same strain, as is often required in metabolic engineering. In addition, we demonstrate the ability to efficiently cure the second plasmid in the system by targeting with Cas9, leaving the cells plasmid-free.
Figures

Similar articles
-
A double-locus scarless genome editing system in Escherichia coli.Biotechnol Lett. 2020 Aug;42(8):1457-1465. doi: 10.1007/s10529-020-02856-7. Epub 2020 Mar 4. Biotechnol Lett. 2020. PMID: 32130564
-
Coupling the CRISPR/Cas9 System with Lambda Red Recombineering Enables Simplified Chromosomal Gene Replacement in Escherichia coli.Appl Environ Microbiol. 2015 Aug;81(15):5103-14. doi: 10.1128/AEM.01248-15. Epub 2015 May 22. Appl Environ Microbiol. 2015. PMID: 26002895 Free PMC article.
-
Deletion of FRT-sites by no-SCAR recombineering in Escherichia coli.Microbiology (Reading). 2022 Apr;168(4). doi: 10.1099/mic.0.001173. Microbiology (Reading). 2022. PMID: 35411846
-
Application of CRISPR/Cas9 genome editing to the study and treatment of disease.Arch Toxicol. 2015 Jul;89(7):1023-34. doi: 10.1007/s00204-015-1504-y. Epub 2015 Apr 1. Arch Toxicol. 2015. PMID: 25827103 Review.
-
Recombineering: genetic engineering in bacteria using homologous recombination.Curr Protoc Mol Biol. 2007 Apr;Chapter 1:Unit 1.16. doi: 10.1002/0471142727.mb0116s78. Curr Protoc Mol Biol. 2007. PMID: 18265390 Review.
Cited by
-
Holistic engineering of cell-free systems through proteome-reprogramming synthetic circuits.Nat Commun. 2020 Jun 19;11(1):3138. doi: 10.1038/s41467-020-16900-7. Nat Commun. 2020. PMID: 32561745 Free PMC article.
-
Generation of bright autobioluminescent bacteria by chromosomal integration of the improved lux operon ilux2.Sci Rep. 2022 Nov 9;12(1):19039. doi: 10.1038/s41598-022-22068-5. Sci Rep. 2022. PMID: 36351939 Free PMC article.
-
Analysing the fitness cost of antibiotic resistance to identify targets for combination antimicrobials.Nat Microbiol. 2021 Nov;6(11):1410-1423. doi: 10.1038/s41564-021-00973-1. Epub 2021 Oct 25. Nat Microbiol. 2021. PMID: 34697460 Free PMC article.
-
A Single Nucleotide Polymorphism in lptG Increases Tolerance to Bile Salts, Acid, and Staining of Calcofluor-Binding Polysaccharides in Salmonella enterica Serovar Typhimurium E40.Front Microbiol. 2021 Jun 2;12:671453. doi: 10.3389/fmicb.2021.671453. eCollection 2021. Front Microbiol. 2021. PMID: 34149657 Free PMC article.
-
A CRISPR-Cas9 Assisted Non-Homologous End-Joining Strategy for One-step Engineering of Bacterial Genome.Sci Rep. 2016 Nov 24;6:37895. doi: 10.1038/srep37895. Sci Rep. 2016. PMID: 27883076 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

