Mutations in the bacterial cell division protein FtsZ highlight the role of GTP binding and longitudinal subunit interactions in assembly and function
- PMID: 26463348
- PMCID: PMC4603965
- DOI: 10.1186/s12866-015-0544-z
Mutations in the bacterial cell division protein FtsZ highlight the role of GTP binding and longitudinal subunit interactions in assembly and function
Abstract
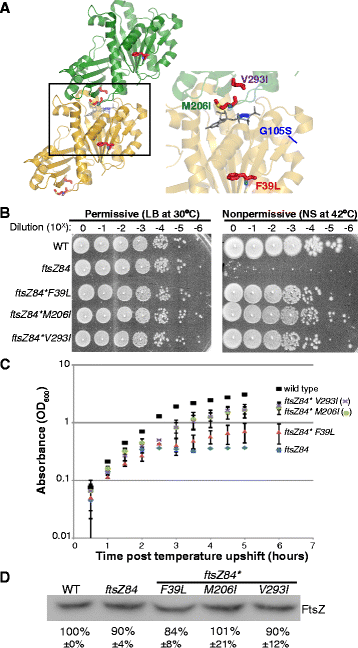
Background: Assembly of the tubulin-like GTPase, FtsZ, at the future division site initiates the process of bacterial cytokinesis. The FtsZ ring serves as a platform for assembly of the division machinery and constricts at the leading edge of the invaginating septum during cytokinesis. In vitro, FtsZ assembles in a GTP-dependent manner, forming straight filaments that curve upon GTP hydrolysis. FtsZ binds but cannot hydrolyze GTP as a monomer. Instead, the active site for GTP hydrolysis is formed at the monomer-monomer interface upon dimerization. While the dynamics of GTP hydrolysis and assembly have been extensively studied in vitro, significantly less is known about the role of GTP binding and hydrolysis in vivo. ftsZ84, a GTPase defective allele of Escherichia coli ftsZ, provides a striking example of the disconnect between in vivo and in vitro FtsZ assembly.
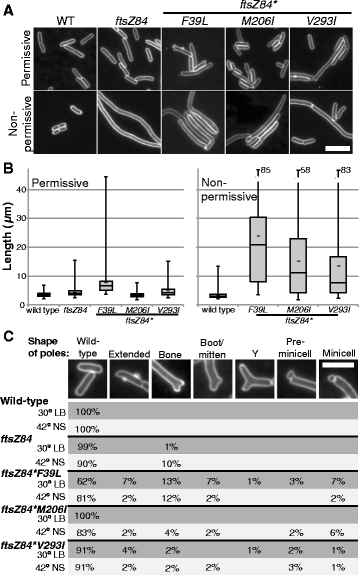
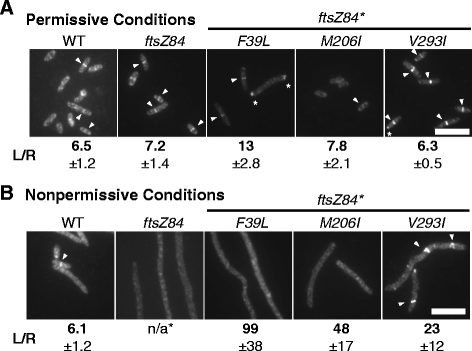
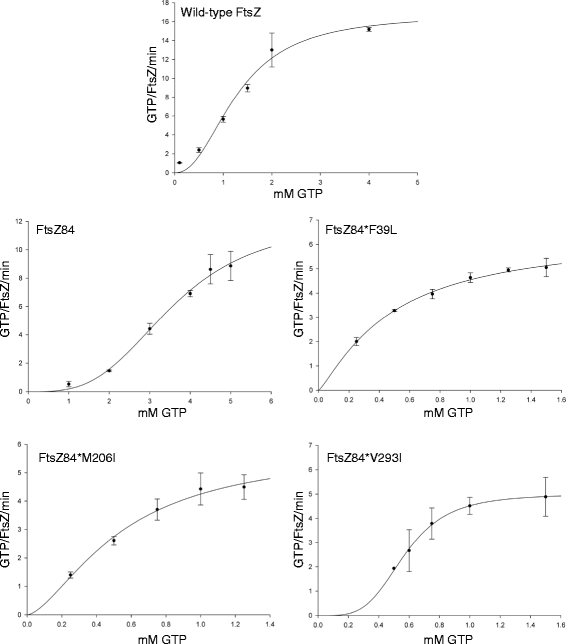
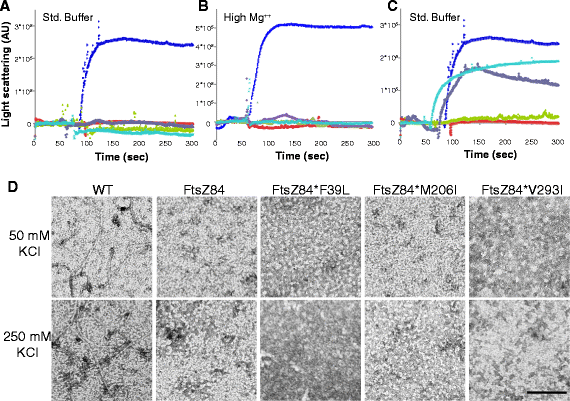
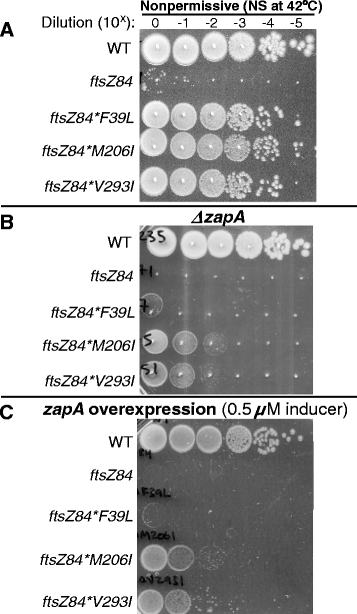
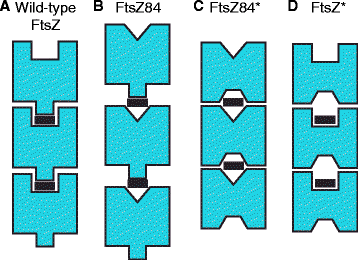
Results: Although ftsZ84 mutants are defective for FtsZ ring formation and division under nonpermissive conditions, they are near wild type for ring formation and division under permissive conditions. In vitro, however, purified FtsZ84 is defective in GTP binding, hydrolysis and assembly under standard reaction conditions. To clarify the nature of the FtsZ84 assembly defect, we isolated and characterized three intragenic suppressors of ftsZ84. All three suppressor mutations increased the apparent affinity of FtsZ84 for GTP, consistent with improved subunit-subunit interactions along the longitudinal interface. Although kinetic analysis indicates that the suppressor mutations increase the affinity of FtsZ84 for GTP, all three exhibit reduced rates of GTP hydrolysis and fail to support assembly in vitro.
Conclusion: Together, our data suggest that FtsZ, and potentially other enzymes whose assembly is similarly regulated, can compensate for defects in catalysis through increases in substrate binding and subunit-subunit interactions. In addition, these results highlight the dichotomy between commonly used in vitro assembly conditions and FtsZ ring formation in the complex intracellular milieu.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

