The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands
- PMID: 26464438
- PMCID: PMC4702778
- DOI: 10.1093/nar/gkv1037
The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands
Abstract
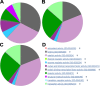
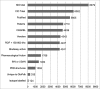
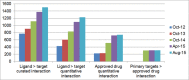
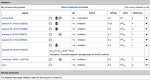
The IUPHAR/BPS Guide to PHARMACOLOGY (GtoPdb, http://www.guidetopharmacology.org) provides expert-curated molecular interactions between successful and potential drugs and their targets in the human genome. Developed by the International Union of Basic and Clinical Pharmacology (IUPHAR) and the British Pharmacological Society (BPS), this resource, and its earlier incarnation as IUPHAR-DB, is described in our 2014 publication. This update incorporates changes over the intervening seven database releases. The unique model of content capture is based on established and new target class subcommittees collaborating with in-house curators. Most information comes from journal articles, but we now also index kinase cross-screening panels. Targets are specified by UniProtKB IDs. Small molecules are defined by PubChem Compound Identifiers (CIDs); ligand capture also includes peptides and clinical antibodies. We have extended the capture of ligands and targets linked via published quantitative binding data (e.g. Ki, IC50 or Kd). The resulting pharmacological relationship network now defines a data-supported druggable genome encompassing 7% of human proteins. The database also provides an expanded substrate for the biennially published compendium, the Concise Guide to PHARMACOLOGY. This article covers content increase, entity analysis, revised curation strategies, new website features and expanded download options.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

