Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus
- PMID: 26464510
- PMCID: PMC4629319
- DOI: 10.1073/pnas.1517952112
Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus
Abstract
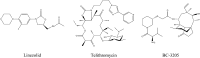
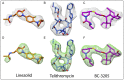
The emergence of bacterial multidrug resistance to antibiotics threatens to cause regression to the preantibiotic era. Here we present the crystal structure of the large ribosomal subunit from Staphylococcus aureus, a versatile Gram-positive aggressive pathogen, and its complexes with the known antibiotics linezolid and telithromycin, as well as with a new, highly potent pleuromutilin derivative, BC-3205. These crystal structures shed light on specific structural motifs of the S. aureus ribosome and the binding modes of the aforementioned antibiotics. Moreover, by analyzing the ribosome structure and comparing it with those of nonpathogenic bacterial models, we identified some unique internal and peripheral structural motifs that may be potential candidates for improving known antibiotics and for use in the design of selective antibiotic drugs against S. aureus.
Keywords: antibiotic resistance; potential advanced pleuoromutilin; protein biosynthesis; species specificity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
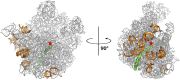


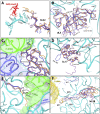
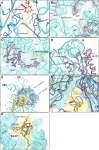




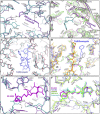

References
-
- Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339(8):520–532. - PubMed
-
- World Health Organization . Antimicrobial Resistance: Global Report on Surveillance. World Health Organization; Geneva: 2014.
-
- Schlünzen F, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase center in eubacteria. Nature. 2001;413(6858):814–821. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

