Auxin signaling modules regulate maize inflorescence architecture
- PMID: 26464512
- PMCID: PMC4629326
- DOI: 10.1073/pnas.1516473112
Auxin signaling modules regulate maize inflorescence architecture
Abstract
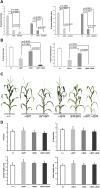
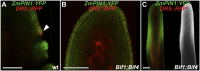
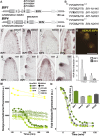
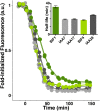
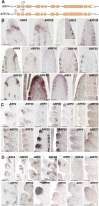
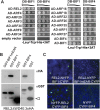
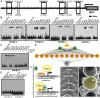
In plants, small groups of pluripotent stem cells called axillary meristems are required for the formation of the branches and flowers that eventually establish shoot architecture and drive reproductive success. To ensure the proper formation of new axillary meristems, the specification of boundary regions is required for coordinating their development. We have identified two maize genes, BARREN INFLORESCENCE1 and BARREN INFLORESCENCE4 (BIF1 and BIF4), that regulate the early steps required for inflorescence formation. BIF1 and BIF4 encode AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) proteins, which are key components of the auxin hormone signaling pathway that is essential for organogenesis. Here we show that BIF1 and BIF4 are integral to auxin signaling modules that dynamically regulate the expression of BARREN STALK1 (BA1), a basic helix-loop-helix (bHLH) transcriptional regulator necessary for axillary meristem formation that shows a striking boundary expression pattern. These findings suggest that auxin signaling directly controls boundary domains during axillary meristem formation and define a fundamental mechanism that regulates inflorescence architecture in one of the most widely grown crop species.
Keywords: auxin signaling; axillary meristems; boundary domains; inflorescence development; maize.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127(7):1309–1321. - PubMed
-
- Bommert P, Nagasawa NS, Jackson D. Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat Genet. 2013;45(3):334–337. - PubMed
-
- Park SJ, et al. Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat Genet. 2014;46(12):1337–1342. - PubMed
-
- Gälweiler L, et al. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282(5397):2226–2230. - PubMed
-
- Przemeck GK, Mattsson J, Hardtke CS, Sung ZR, Berleth T. Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta. 1996;200(2):229–237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

