Notch signal strength controls cell fate in the haemogenic endothelium
- PMID: 26465397
- PMCID: PMC4634136
- DOI: 10.1038/ncomms9510
Notch signal strength controls cell fate in the haemogenic endothelium
Erratum in
-
Corrigendum: Notch signal strength controls cell fate in the haemogenic endothelium.Nat Commun. 2016 Mar 4;7:10978. doi: 10.1038/ncomms10978. Nat Commun. 2016. PMID: 26939771 Free PMC article. No abstract available.
Abstract
Acquisition of the arterial and haemogenic endothelium fates concurrently occur in the aorta-gonad-mesonephros (AGM) region prior to haematopoietic stem cell (HSC) generation. The arterial programme depends on Dll4 and the haemogenic endothelium/HSC on Jag1-mediated Notch1 signalling. How Notch1 distinguishes and executes these different programmes in response to particular ligands is poorly understood. By using two Notch1 activation trap mouse models with different sensitivity, here we show that arterial endothelial cells and HSCs originate from distinct precursors, characterized by different Notch1 signal strengths. Microarray analysis on AGM subpopulations demonstrates that the Jag1 ligand stimulates low Notch strength, inhibits the endothelial programme and is permissive for HSC specification. In the absence of Jag1, endothelial cells experience high Dll4-induced Notch activity and select the endothelial programme, thus precluding HSC formation. Interference with the Dll4 signal by ligand-specific blocking antibodies is sufficient to inhibit the endothelial programme and favour specification of the haematopoietic lineage.
Conflict of interest statement
Minhong Yan is an employee and shareholder of Roche Genentech. The remaining authors declare no competing financial interest.
Figures
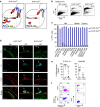
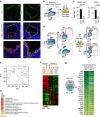
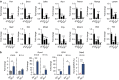
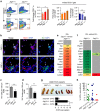


References
-
- Medvinsky A. & Dzierzak E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell 86, 897–906 (1996). - PubMed
-
- Kissa K. & Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464, 112–115 (2010). - PubMed
-
- Boisset J. C. et al. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

