Ribosome profiling reveals the what, when, where and how of protein synthesis
- PMID: 26465719
- PMCID: PMC5522010
- DOI: 10.1038/nrm4069
Ribosome profiling reveals the what, when, where and how of protein synthesis
Abstract
Ribosome profiling, which involves the deep sequencing of ribosome-protected mRNA fragments, is a powerful tool for globally monitoring translation in vivo. The method has facilitated discovery of the regulation of gene expression underlying diverse and complex biological processes, of important aspects of the mechanism of protein synthesis, and even of new proteins, by providing a systematic approach for experimental annotation of coding regions. Here, we introduce the methodology of ribosome profiling and discuss examples in which this approach has been a key factor in guiding biological discovery, including its prominent role in identifying thousands of novel translated short open reading frames and alternative translation products.
Conflict of interest statement
Figures

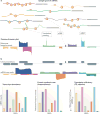
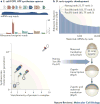
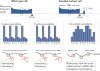
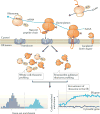

References
-
- Ellis SR. Nucleolar stress in Diamond Blackfan anemia pathophysiology. Biochim Biophys Acta. 2014;1842:765–768. - PubMed

