Mutations driving CLL and their evolution in progression and relapse
- PMID: 26466571
- PMCID: PMC4815041
- DOI: 10.1038/nature15395
Mutations driving CLL and their evolution in progression and relapse
Abstract
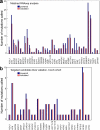
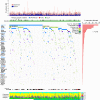
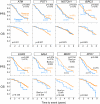
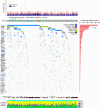
Which genetic alterations drive tumorigenesis and how they evolve over the course of disease and therapy are central questions in cancer biology. Here we identify 44 recurrently mutated genes and 11 recurrent somatic copy number variations through whole-exome sequencing of 538 chronic lymphocytic leukaemia (CLL) and matched germline DNA samples, 278 of which were collected in a prospective clinical trial. These include previously unrecognized putative cancer drivers (RPS15, IKZF3), and collectively identify RNA processing and export, MYC activity, and MAPK signalling as central pathways involved in CLL. Clonality analysis of this large data set further enabled reconstruction of temporal relationships between driver events. Direct comparison between matched pre-treatment and relapse samples from 59 patients demonstrated highly frequent clonal evolution. Thus, large sequencing data sets of clinically informative samples enable the discovery of novel genes associated with cancer, the network of relationships between the driver events, and their impact on disease relapse and clinical outcome.
Figures









Comment in
-
Genetic profiling of CLL: a 'TP53 addict' perspective.Cell Death Dis. 2016 Jan 14;7(1):e2042. doi: 10.1038/cddis.2015.415. Cell Death Dis. 2016. PMID: 26775696 Free PMC article. No abstract available.
References
-
- Quesada V, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012;44:47–52. doi:10.1038/ng.1032. - PubMed
-
- Schuh A, et al. Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood. 2012;120:4191–4196. doi:10.1182/blood-2012-05-433540. - PubMed
Additional bibliography for Extended Data Figures
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- U10 CA180861/CA/NCI NIH HHS/United States
- 1U10CA180861-01/CA/NCI NIH HHS/United States
- R01 CA182461/CA/NCI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- 1K01ES025431-01/ES/NIEHS NIH HHS/United States
- 1R01CA182461-02/CA/NCI NIH HHS/United States
- U54HG003067/HG/NHGRI NIH HHS/United States
- K01 ES025431/ES/NIEHS NIH HHS/United States
- P01 CA081534/CA/NCI NIH HHS/United States
- 1R01CA184922-01/CA/NCI NIH HHS/United States
- R01 CA184922/CA/NCI NIH HHS/United States
- R01 HL116452/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

