Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species
- PMID: 26467257
- PMCID: PMC5057366
- DOI: 10.1111/nph.13697
Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species
Abstract
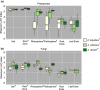
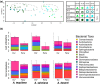
Desert plants are hypothesized to survive the environmental stress inherent to these regions in part thanks to symbioses with microorganisms, and yet these microbial species, the communities they form, and the forces that influence them are poorly understood. Here we report the first comprehensive investigation of the microbial communities associated with species of Agave, which are native to semiarid and arid regions of Central and North America and are emerging as biofuel feedstocks. We examined prokaryotic and fungal communities in the rhizosphere, phyllosphere, leaf and root endosphere, as well as proximal and distal soil samples from cultivated and native agaves, through Illumina amplicon sequencing. Phylogenetic profiling revealed that the composition of prokaryotic communities was primarily determined by the plant compartment, whereas the composition of fungal communities was mainly influenced by the biogeography of the host species. Cultivated A. tequilana exhibited lower levels of prokaryotic diversity compared with native agaves, although no differences in microbial diversity were found in the endosphere. Agaves shared core prokaryotic and fungal taxa known to promote plant growth and confer tolerance to abiotic stress, which suggests common principles underpinning Agave-microbe interactions.
Keywords: Agave; biogeography; cultivation; desert; iTags; microbial diversity; plant microbiome; plant-microbe interactions.
No claim to US Government works. New Phytologist © 2015 New Phytologist Trust.
Figures




Comment in
-
Disentangling the factors shaping microbiota composition across the plant holobiont.New Phytol. 2016 Jan;209(2):454-7. doi: 10.1111/nph.13760. New Phytol. 2016. PMID: 26763678 No abstract available.
References
-
- Borland AM, Griffiths H, Hartwell J, Smith JAC. 2009. Exploiting the potential of plants with crassulacean acid metabolism for bioenergy production on marginal lands. Journal of Experimental Botany 60: 2879–2896. - PubMed
-
- Bulgarelli D, Rott M, Schlaeppi K, Loren Ver, van Themaat E, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R et al 2012. Revealing structure and assembly cues for Arabidopsis root‐inhabiting bacterial microbiota. Nature 488: 91–95. - PubMed
-
- Campos H, Trejo C, Peña‐Valdivia CB, García‐Nava R, Conde‐Martínez FV, Cruz‐Ortega Mdel R. 2014. Photosynthetic acclimation to drought stress in Agave salmiana Otto ex Salm‐Dyck seedlings is largely dependent on thermal dissipation and enhanced electron flux to photosystem I. Photosynthesis Research 122: 1–17. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

