ChEBI in 2016: Improved services and an expanding collection of metabolites
- PMID: 26467479
- PMCID: PMC4702775
- DOI: 10.1093/nar/gkv1031
ChEBI in 2016: Improved services and an expanding collection of metabolites
Abstract
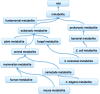
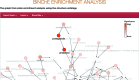
ChEBI is a database and ontology containing information about chemical entities of biological interest. It currently includes over 46,000 entries, each of which is classified within the ontology and assigned multiple annotations including (where relevant) a chemical structure, database cross-references, synonyms and literature citations. All content is freely available and can be accessed online at http://www.ebi.ac.uk/chebi. In this update paper, we describe recent improvements and additions to the ChEBI offering. We have substantially extended our collection of endogenous metabolites for several organisms including human, mouse, Escherichia coli and yeast. Our front-end has also been reworked and updated, improving the user experience, removing our dependency on Java applets in favour of embedded JavaScript components and moving from a monthly release update to a 'live' website. Programmatic access has been improved by the introduction of a library, libChEBI, in Java, Python and Matlab. Furthermore, we have added two new tools, namely an analysis tool, BiNChE, and a query tool for the ontology, OntoQuery.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

