Multipotent luminal mammary cancer stem cells model tumor heterogeneity
- PMID: 26467658
- PMCID: PMC4606989
- DOI: 10.1186/s13058-015-0615-y
Multipotent luminal mammary cancer stem cells model tumor heterogeneity
Abstract
Introduction: The diversity of human breast cancer subtypes has led to the hypothesis that breast cancer is actually a number of different diseases arising from cells at various stages of differentiation. The elusive nature of the cell(s) of origin thus hampers approaches to eradicate the disease.
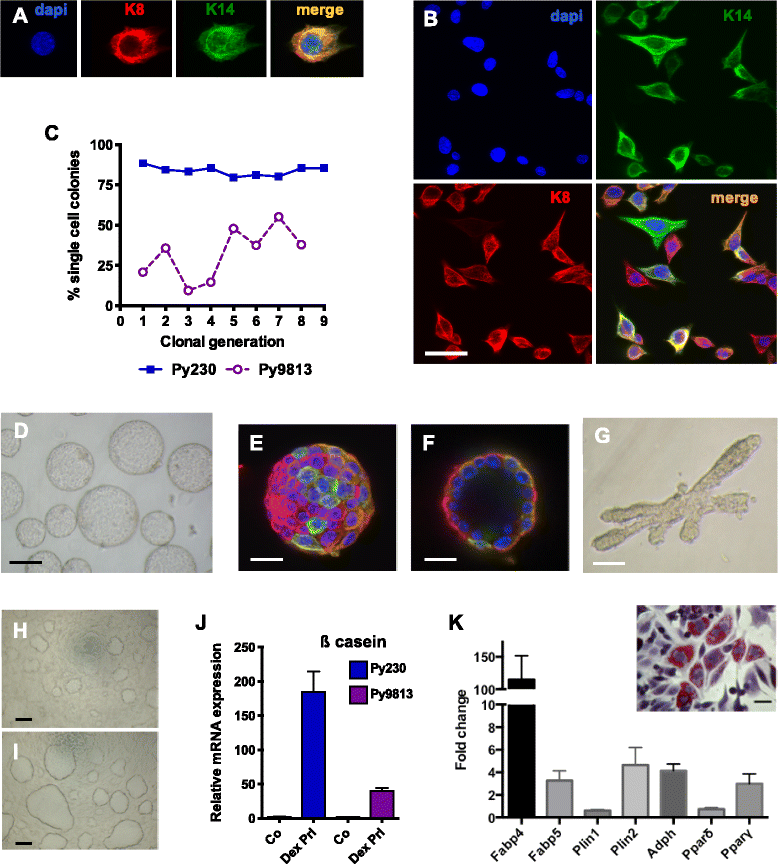
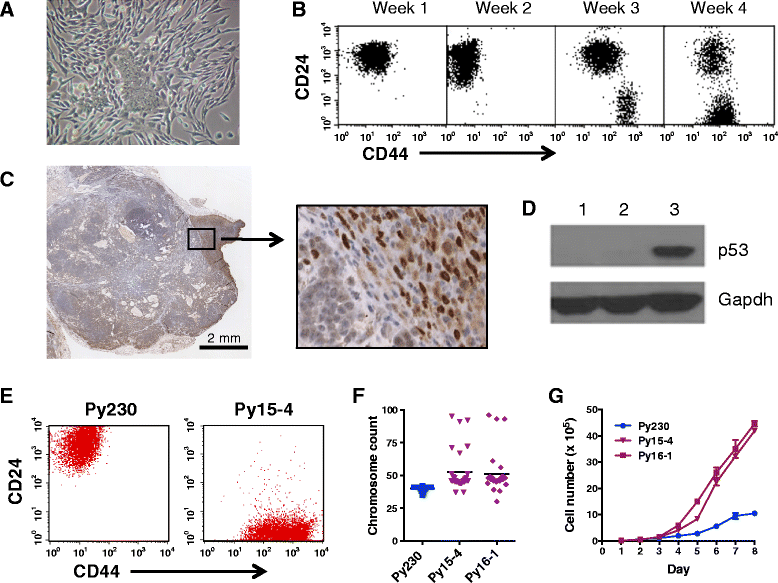
Methods: Clonal cell lines were isolated from primary transgenic polyomavirus middle T (PyVmT) luminal tumors. Mammary cancer stem cell (MaCSC) properties were examined by immunofluorescence, flow cytometry, differentiation assays and in vivo tumorigenesis.
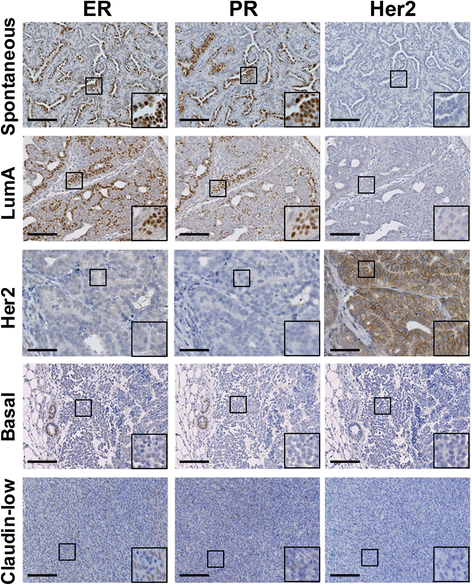
Results: Clonal cell lines isolated from primary PyVmT mouse mammary luminal tumors can differentiate into luminal, myoepithelial, alveolar and adipocyte lineages. Upon orthotopic injection, progeny of a single cell follow a pattern of progression from ductal carcinoma in situ, to adenoma, adenocarcinoma and epithelial metastasis that recapitulates the transgenic model. Tumors can evolve in vivo from hormone receptor-positive to hormone receptor-negative Her2-positive, or triple negative CD44hi basal-like and claudin-low tumors. Contrary to the current paradigm, we have defined a model in which multiple tumor subtypes can originate from a single multipotent cancer stem cell that undergoes genetic and/or epigenetic evolution during tumor progression. As in human tumors, the more aggressive tumor subtypes express nuclear p53. Tumor cell lines can also be derived from these more advanced tumor subtypes.
Conclusions: Since the majority of human tumors are of the luminal subtype, understanding the cell of origin of these tumors and how they relate to other tumor subtypes will impact cancer therapy. Analysis of clonal cell lines derived from different tumor subtypes suggests a developmental hierarchy of MaCSCs, which may provide insights into the progression of human breast cancer.
Figures



References
-
- Wellings S, Jensen M, Marcum R. An atlas of subgross pathology of the human breast with special reference to possible precancerous lesions. J Natl Cancer Inst. 1975;55:231–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

