Genomic Views of Transcriptional Enhancers: Essential Determinants of Cellular Identity and Activity-Dependent Responses in the CNS
- PMID: 26468181
- PMCID: PMC4604220
- DOI: 10.1523/JNEUROSCI.2622-15.2015
Genomic Views of Transcriptional Enhancers: Essential Determinants of Cellular Identity and Activity-Dependent Responses in the CNS
Abstract
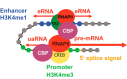
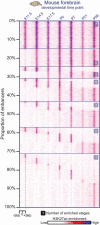
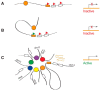
Sprinkled throughout the genome are a million regulatory sequences called transcriptional enhancers that activate gene promoters in the right cells, at the right time. Enhancers endow the brain with its incredible diversity of cell types and also translate neural activity into gene induction. Thanks to rapid advances in genomic technologies, it is now possible to identify thousands of enhancers rapidly, test their transcriptional function en masse, and address their neurobiological functions via genome editing. Enhancers also promise to be a great technological opportunity for neuroscience, offering the potential for cell-type-specific genetic labeling and manipulation without the need for transgenesis. The objective of this review and the accompanying 2015 SfN mini-symposium is to highlight the use of new and emerging genomic technologies to probe enhancer function in the nervous system.
Significance statement: Transcriptional enhancers turn on genes in the right cells, at the right time. Enhancers are also the genomic sequences that encode the incredible diversity of cell types in the brain and enable the brain to turn genes on in response to new experiences. New technology enables enhancers to be found and manipulated. The study of enhancers promises to inform our understanding of brain development and function. The application of enhancer technology holds promise in accelerating basic neuroscience research and enabling gene therapies to be targeted to specific cell types in the brain.
Copyright © 2015 the authors 0270-6474/15/3513819-08$15.00/0.
Figures
Similar articles
-
Decoding enhancers using massively parallel reporter assays.Genomics. 2015 Sep;106(3):159-164. doi: 10.1016/j.ygeno.2015.06.005. Epub 2015 Jun 10. Genomics. 2015. PMID: 26072433 Free PMC article. Review.
-
Long-range enhancer-promoter contacts in gene expression control.Nat Rev Genet. 2019 Aug;20(8):437-455. doi: 10.1038/s41576-019-0128-0. Nat Rev Genet. 2019. PMID: 31086298 Review.
-
Genomic Enhancers in Brain Health and Disease.Genes (Basel). 2019 Jan 14;10(1):43. doi: 10.3390/genes10010043. Genes (Basel). 2019. PMID: 30646598 Free PMC article. Review.
-
Transcriptional enhancers and their communication with gene promoters.Cell Mol Life Sci. 2021 Oct;78(19-20):6453-6485. doi: 10.1007/s00018-021-03903-w. Epub 2021 Aug 19. Cell Mol Life Sci. 2021. PMID: 34414474 Free PMC article. Review.
-
Neurobiological functions of transcriptional enhancers.Nat Neurosci. 2020 Jan;23(1):5-14. doi: 10.1038/s41593-019-0538-5. Epub 2019 Nov 18. Nat Neurosci. 2020. PMID: 31740812 Free PMC article. Review.
Cited by
-
The chromatin landscape of neuronal plasticity.Curr Opin Neurobiol. 2019 Dec;59:79-86. doi: 10.1016/j.conb.2019.04.006. Epub 2019 Jun 4. Curr Opin Neurobiol. 2019. PMID: 31174107 Free PMC article. Review.
-
Invited Review: Epigenetics in neurodevelopment.Neuropathol Appl Neurobiol. 2020 Feb;46(1):6-27. doi: 10.1111/nan.12608. Epub 2020 Mar 9. Neuropathol Appl Neurobiol. 2020. PMID: 32056273 Free PMC article. Review.
-
Maximizing lentiviral vector gene transfer in the CNS.Gene Ther. 2021 Feb;28(1-2):75-88. doi: 10.1038/s41434-020-0172-6. Epub 2020 Jul 6. Gene Ther. 2021. PMID: 32632267 Free PMC article.
-
Chromatin Regulation of Neuronal Maturation and Plasticity.Trends Neurosci. 2018 May;41(5):311-324. doi: 10.1016/j.tins.2018.02.009. Epub 2018 Mar 9. Trends Neurosci. 2018. PMID: 29530320 Free PMC article. Review.
-
Negative Evidence for a Functional Role of Neuronal DNMT3a in Persistent Pain.Front Mol Neurosci. 2018 Sep 12;11:332. doi: 10.3389/fnmol.2018.00332. eCollection 2018. Front Mol Neurosci. 2018. PMID: 30258352 Free PMC article.
References
-
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. - DOI - PMC - PubMed
-
- Andzelm MM, Cherry TJ, Harmin DA, Boeke AC, Lee C, Hemberg M, Pawlyk B, Malik AN, Flavell SW, Sandberg MA, Raviola E, Greenberg ME. MEF2D drives photoreceptor development through a genome-wide competition for tissue-specific enhancers. Neuron. 2015;86:247–263. doi: 10.1016/j.neuron.2015.02.038. - DOI - PMC - PubMed
-
- Arloth J, Bogdan R, Weber P, Frishman G, Menke A, Wagner KV, Balsevich G, Schmidt MV, Karbalai N, Czamara D, Altmann A, Trümbach D, Wurst W, Mehta D, Uhr M, Klengel T, Erhardt A, Carey CE, Conley ED, et al. Genetic differences in the immediate transcriptome response to stress predict risk-related brain function and psychiatric disorders. Neuron. 2015;86:1189–1202. doi: 10.1016/j.neuron.2015.05.034. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
