Reducing Ribosomal Protein S6 Kinase 1 Expression Improves Spatial Memory and Synaptic Plasticity in a Mouse Model of Alzheimer's Disease
- PMID: 26468204
- PMCID: PMC4604237
- DOI: 10.1523/JNEUROSCI.2781-15.2015
Reducing Ribosomal Protein S6 Kinase 1 Expression Improves Spatial Memory and Synaptic Plasticity in a Mouse Model of Alzheimer's Disease
Abstract
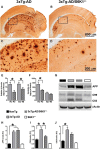
Aging is the most important risk factor associated with Alzheimer's disease (AD); however, the molecular mechanisms linking aging to AD remain unclear. Suppression of the ribosomal protein S6 kinase 1 (S6K1) increases healthspan and lifespan in several organisms, from nematodes to mammals. Here we show that S6K1 expression is upregulated in the brains of AD patients. Using a mouse model of AD, we found that genetic reduction of S6K1 improved synaptic plasticity and spatial memory deficits, and reduced the accumulation of amyloid-β and tau, the two neuropathological hallmarks of AD. Mechanistically, these changes were linked to reduced translation of tau and the β-site amyloid precursor protein cleaving enzyme 1, a key enzyme in the generation of amyloid-β. Our results implicate S6K1 dysregulation as a previously unidentified molecular mechanism underlying synaptic and memory deficits in AD. These findings further suggest that therapeutic manipulation of S6K1 could be a valid approach to mitigate AD pathology.
Significance statement: Aging is the most important risk factor for Alzheimer's disease (AD). However, little is known about how it contributes to AD pathogenesis. S6 kinase 1 (S6K1) is a protein kinase involved in regulation of protein translation. Reducing S6K1 activity increases lifespan and healthspan. We report the novel finding that reducing S6K1 activity in 3xTg-AD mice ameliorates synaptic and cognitive deficits. These improvement were associated with a reduction in amyloid-β and tau pathology. Mechanistically, lowering S6K1 levels reduced translation of β-site amyloid precursor protein cleaving enzyme 1 and tau, two key proteins involved in AD pathogenesis. These data suggest that S6K1 may represent a molecular link between aging and AD. Given that aging is the most important risk factor for most neurodegenerative diseases, our results may have far-reaching implications into other diseases.
Keywords: AD; Aβ; aging; mTOR; plaques; tangles.
Copyright © 2015 the authors 0270-6474/15/3514042-15$15.00/0.
Figures







References
-
- Alzheimer's Association. 2014 Alzheimer's disease facts and figures. Alzheimer's Dement. 2014;10:e47–e92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
