Subunits of ADA-two-A-containing (ATAC) or Spt-Ada-Gcn5-acetyltrasferase (SAGA) Coactivator Complexes Enhance the Acetyltransferase Activity of GCN5
- PMID: 26468280
- PMCID: PMC4661412
- DOI: 10.1074/jbc.M115.668533
Subunits of ADA-two-A-containing (ATAC) or Spt-Ada-Gcn5-acetyltrasferase (SAGA) Coactivator Complexes Enhance the Acetyltransferase Activity of GCN5
Abstract
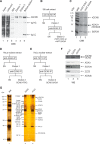
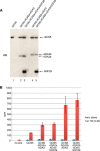
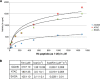
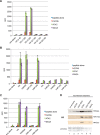
Histone acetyl transferases (HATs) play a crucial role in eukaryotes by regulating chromatin architecture and locus specific transcription. GCN5 (KAT2A) is a member of the GNAT (Gcn5-related N-acetyltransferase) family of HATs. In metazoans this enzyme is found in two functionally distinct coactivator complexes, SAGA (Spt Ada Gcn5 acetyltransferase) and ATAC (Ada Two A-containing). These two multiprotein complexes comprise complex-specific and shared subunits, which are organized in functional modules. The HAT module of ATAC is composed of GCN5, ADA2a, ADA3, and SGF29, whereas in the SAGA HAT module ADA2b is present instead of ADA2a. To better understand how the activity of human (h) hGCN5 is regulated in the two related, but different, HAT complexes we carried out in vitro HAT assays. We compared the activity of hGCN5 alone with its activity when it was part of purified recombinant hATAC or hSAGA HAT modules or endogenous hATAC or hSAGA complexes using histone tail peptides and full-length histones as substrates. We demonstrated that the subunit environment of the HAT complexes into which GCN5 incorporates determines the enhancement of GCN5 activity. On histone peptides we show that all the tested GCN5-containing complexes acetylate mainly histone H3K14. Our results suggest a stronger influence of ADA2b as compared with ADA2a on the activity of GCN5. However, the lysine acetylation specificity of GCN5 on histone tails or full-length histones was not changed when incorporated in the HAT modules of ATAC or SAGA complexes. Our results thus demonstrate that the catalytic activity of GCN5 is stimulated by subunits of the ADA2a- or ADA2b-containing HAT modules and is further increased by incorporation of the distinct HAT modules in the ATAC or SAGA holo-complexes.
Keywords: KAT2A; chromatin; chromatin modification; chromatin regulation; complex; histone; histone acetylase; histone acetylation; human; mass spectrometry (MS).
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
-
- Kornberg R. D., and Lorch Y. (1999) Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98, 285–294 - PubMed
-
- Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 - PubMed
-
- Hong L., Schroth G. P., Matthews H. R., Yau P., and Bradbury E. M. (1993) Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J. Biol. Chem. 268, 305–314 - PubMed
-
- Li R., Yang L., Fouts E., and Botchan M. R. (1993) Site-specific DNA-binding proteins important for replication and transcription have multiple activities. Cold Spring Harb. Symp. Quant. Biol. 58, 403–413 - PubMed
-
- Nagy Z., and Tora L. (2007) Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 26, 5341–5357 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

