Sequence Variation in Amplification Target Genes and Standards Influences Interlaboratory Comparison of BK Virus DNA Load Measurement
- PMID: 26468499
- PMCID: PMC4652096
- DOI: 10.1128/JCM.02145-15
Sequence Variation in Amplification Target Genes and Standards Influences Interlaboratory Comparison of BK Virus DNA Load Measurement
Abstract
International guidelines define a BK virus (BKV) load of ≥4 log10 copies/ml as presumptive of BKV-associated nephropathy (BKVN) and a cutoff for therapeutic intervention. To investigate whether BKV DNA loads (BKVL) are comparable between laboratories, 2 panels of 15 and 8 clinical specimens (urine, whole blood, and plasma) harboring different BKV genotypes were distributed to 20 and 27 French hospital centers in 2013 and 2014, respectively. Although 68% of the reported results fell within the acceptable range of the expected result ±0.5 log10, the interlaboratory variation ranged from 1.32 to 5.55 log10. Polymorphisms specific to BKV genotypes II and IV, namely, the number and position of mutations in amplification target genes and/or deletion in standards, arose as major sources of interlaboratory disagreements. The diversity of DNA purification methods also contributed to the interlaboratory variability, in particular for urine samples. Our data strongly suggest that (i) commercial external quality controls for BKVL assessment should include all major BKV genotypes to allow a correct evaluation of BKV assays, and (ii) the BKV sequence of commercial standards should be provided to users to verify the absence of mismatches with the primers and probes of their BKV assays. Finally, the optimization of primer and probe design and standardization of DNA extraction methods may substantially decrease interlaboratory variability and allow interinstitutional studies to define a universal cutoff for presumptive BKVN and, ultimately, ensure adequate patient care.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
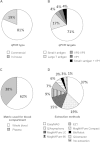



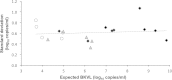
References
-
- Hirsch HH, Vincenti F, Friman S, Tuncer M, Citterio F, Wiecek A, Scheuermann EH, Klinger M, Russ G, Pescovitz MD, Prestele H. 2013. Polyomavirus BK replication in de novo kidney transplant patients receiving tacrolimus or cyclosporine: a prospective, randomized, multicenter study. Am J Transplant 13:136–145. doi:10.1111/j.1600-6143.2012.04320.x. - DOI - PMC - PubMed
-
- Drachenberg RC, Drachenberg CB, PapadimitriouC J, Ramos E, Fink JC, Wali R, Weir MR, Cangro CB, Klassen DK, Khaled A, Cunningham R, Bartlett ST. 2001. Morphological spectrum of polyoma virus disease in renal allografts: diagnostic accuracy of urine cytology. Am J Transplant 1:373–381. doi:10.1034/j.1600-6143.2001.10414.x. - DOI - PubMed

