pH-Dependent Formation and Disintegration of the Influenza A Virus Protein Scaffold To Provide Tension for Membrane Fusion
- PMID: 26468548
- PMCID: PMC4702562
- DOI: 10.1128/JVI.01539-15
pH-Dependent Formation and Disintegration of the Influenza A Virus Protein Scaffold To Provide Tension for Membrane Fusion
Abstract
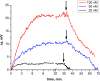
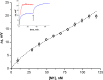
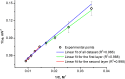
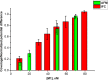
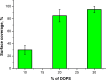
Influenza virus is taken up from a pH-neutral extracellular milieu into an endosome, whose contents then acidify, causing changes in the viral matrix protein (M1) that coats the inner monolayer of the viral lipid envelope. At a pH of ~6, M1 interacts with the viral ribonucleoprotein (RNP) in a putative priming stage; at this stage, the interactions of the M1 scaffold coating the lipid envelope are intact. The M1 coat disintegrates as acidification continues to a pH of ~5 to clear a physical path for the viral genome to transit from the viral interior to the cytoplasm. Here we investigated the physicochemical mechanism of M1's pH-dependent disintegration. In neutral media, the adsorption of M1 protein on the lipid bilayer was electrostatic in nature and reversible. The energy of the interaction of M1 molecules with each other in M1 dimers was about 10 times as weak as that of the interaction of M1 molecules with the lipid bilayer. Acidification drives conformational changes in M1 molecules due to changes in the M1 charge, leading to alterations in their electrostatic interactions. Dropping the pH from 7.1 to 6.0 did not disturb the M1 layer; dropping it lower partially desorbed M1 because of increased repulsion between M1 monomers still stuck to the membrane. Lipid vesicles coated with M1 demonstrated pH-dependent rupture of the vesicle membrane, presumably because of the tension generated by this repulsive force. Thus, the disruption of the vesicles coincident with M1 protein scaffold disintegration at pH 5 likely stretches the lipid membrane to the point of rupture, promoting fusion pore widening for RNP release.
Importance: Influenza remains a top killer of human beings throughout the world, in part because of the influenza virus's rapid binding to cells and its uptake into compartments hidden from the immune system. To attack the influenza virus during this time of hiding, we need to understand the physical forces that allow the internalized virus to infect the cell. In particular, we need to know how the protective coat of protein inside the viral surface reacts to the changes in acid that come soon after internalization. We found that acid makes the molecules of the protein coat push each other while they are still stuck to the virus, so that they would like to rip the membrane apart. This ripping force is known to promote membrane fusion, the process by which infection actually occurs.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures






References
-
- Lamb RA, Krug RM. 2001. Orthomyxoviridae: the viruses and their replication, p 1487–1532. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Sun X, Whittaker GR. 2006. Entry of influenza virus, p 72–82. In Poehlmann S, Simmons G (ed), Viral entry into host cells. Springer Science & Business Media, LLC, New York, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

