Small RNA Sequencing Uncovers New miRNAs and moRNAs Differentially Expressed in Normal and Primary Myelofibrosis CD34+ Cells
- PMID: 26468945
- PMCID: PMC4607157
- DOI: 10.1371/journal.pone.0140445
Small RNA Sequencing Uncovers New miRNAs and moRNAs Differentially Expressed in Normal and Primary Myelofibrosis CD34+ Cells
Abstract
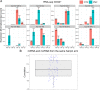
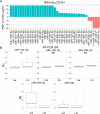
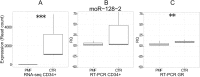
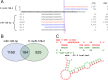
Myeloproliferative neoplasms (MPN) are chronic myeloid cancers thought to arise at the level of CD34+ hematopoietic stem/progenitor cells. They include essential thrombocythemia (ET), polycythemia vera (PV) and primary myelofibrosis (PMF). All can progress to acute leukemia, but PMF carries the worst prognosis. Increasing evidences indicate that deregulation of microRNAs (miRNAs) might plays an important role in hematologic malignancies, including MPN. To attain deeper knowledge of short RNAs (sRNAs) expression pattern in CD34+ cells and of their possible role in mediating post-transcriptional regulation in PMF, we sequenced with Illumina HiSeq2000 technology CD34+ cells from healthy subjects and PMF patients. We detected the expression of 784 known miRNAs, with a prevalence of miRNA up-regulation in PMF samples, and discovered 34 new miRNAs and 99 new miRNA-offset RNAs (moRNAs), in CD34+ cells. Thirty-seven small RNAs were differentially expressed in PMF patients compared with healthy subjects, according to microRNA sequencing data. Five miRNAs (miR-10b-5p, miR-19b-3p, miR-29a-3p, miR-379-5p, and miR-543) were deregulated also in PMF granulocytes. Moreover, 3'-moR-128-2 resulted consistently downregulated in PMF according to RNA-seq and qRT-PCR data both in CD34+ cells and granulocytes. Target predictions of these validated small RNAs de-regulated in PMF and functional enrichment analyses highlighted many interesting pathways involved in tumor development and progression, such as signaling by FGFR and DAP12 and Oncogene Induced Senescence. As a whole, data obtained in this study deepened the knowledge of miRNAs and moRNAs altered expression in PMF CD34+ cells and allowed to identify and validate a specific small RNA profile that distinguishes PMF granulocytes from those of normal subjects. We thus provided new information regarding the possible role of miRNAs and, specifically, of new moRNAs in this disease.
Conflict of interest statement
Figures


References
-
- Tefferi A, Vardiman JW. Classification and diagnosis of myeloproliferative neoplasms: The 2008 World Health Organization criteria and point-of-care diagnostic algorithms. Leukemia. 2007;22: 14–22. - PubMed
-
- Vannucchi AM, Guglielmelli P, Tefferi A. Advances in Understanding and Management of Myeloproliferative Neoplasms. 2009;59: 171–191. - PubMed
-
- Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. 2005;365: 1054–1061. - PubMed
-
- Jones AV, Kreil S, Zoi K, Waghorn K, Curtis C, Zhang L, et al. Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood. 2005;106: 2162–2168. - PubMed
-
- James C, Ugo V, Le Couédic J, Staerk J, Delhommeau F, Lacout C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434: 1144–1148. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

