Single-cell RNA-Seq resolves cellular complexity in sensory organs from the neonatal inner ear
- PMID: 26469390
- PMCID: PMC4634134
- DOI: 10.1038/ncomms9557
Single-cell RNA-Seq resolves cellular complexity in sensory organs from the neonatal inner ear
Abstract
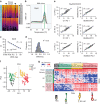
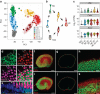
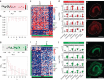
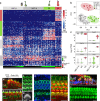
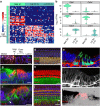
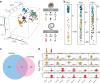
In the inner ear, cochlear and vestibular sensory epithelia utilize grossly similar cell types to transduce different stimuli: sound and acceleration. Each individual sensory epithelium is composed of highly heterogeneous populations of cells based on physiological and anatomical criteria. However, limited numbers of each cell type have impeded transcriptional characterization. Here we generated transcriptomes for 301 single cells from the utricular and cochlear sensory epithelia of newborn mice to circumvent this challenge. Cluster analysis indicates distinct profiles for each of the major sensory epithelial cell types, as well as less-distinct sub-populations. Asynchrony within utricles allows reconstruction of the temporal progression of cell-type-specific differentiation and suggests possible plasticity among cells at the sensory-nonsensory boundary. Comparisons of cell types from utricles and cochleae demonstrate divergence between auditory and vestibular cells, despite a common origin. These results provide significant insights into the developmental processes that form unique inner ear cell types.
Figures



References
-
- Eatock R. A. & Songer J. E. Vestibular hair cells and afferents: two channels for head motion signals. Annu. Rev. Neurosci. 34, 501–534 (2011). - PubMed
-
- Kelley M. W. Regulation of cell fate in the sensory epithelia of the inner ear. Nat. Rev. Neurosci. 7, 837–849 (2006). - PubMed
-
- Fettiplace R. & Hackney C. M. The sensory and motor roles of auditory hair cells. Nat. Rev. Neurosci. 7, 19–29 (2006). - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

