Identification of novel sRNAs involved in biofilm formation, motility, and fimbriae formation in Escherichia coli
- PMID: 26469694
- PMCID: PMC4606813
- DOI: 10.1038/srep15287
Identification of novel sRNAs involved in biofilm formation, motility, and fimbriae formation in Escherichia coli
Abstract
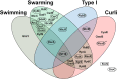
Bacterial small RNAs (sRNAs) are known regulators in many physiological processes. In Escherichia coli, a large number of sRNAs have been predicted, among which only about a hundred are experimentally validated. Despite considerable research, the majority of their functions remain uncovered. Therefore, collective analysis of the roles of sRNAs in specific cellular processes may provide an effective approach to identify their functions. Here, we constructed a collection of plasmids overexpressing 99 individual sRNAs, and analyzed their effects on biofilm formation and related phenotypes. Thirty-three sRNAs significantly affecting these cellular processes were identified. No consistent correlations were observed, except that all five sRNAs suppressing type I fimbriae inhibited biofilm formation. Interestingly, IS118, yet to be characterized, suppressed all the processes. Our data not only reveal potentially critical functions of individual sRNAs in biofilm formation and other phenotypes but also highlight the unexpected complexity of sRNA-mediated metabolic pathways leading to these processes.
Figures









References
-
- Rivas E., Klein R. J., Jones T. A. & Eddy S. R. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr Biol. 11, 1369–1373 (2001). - PubMed
-
- Argaman L. et al. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 11, 941–950 (2001). - PubMed
-
- Chen S. et al. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Biosystems. 65, 157–177 (2002). - PubMed
-
- Zhang A. et al. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol. 50, 1111–1124 (2003). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

