Scalable and cost-effective NGS genotyping in the cloud
- PMID: 26470712
- PMCID: PMC4608296
- DOI: 10.1186/s12920-015-0134-9
Scalable and cost-effective NGS genotyping in the cloud
Abstract
Background: While next-generation sequencing (NGS) costs have plummeted in recent years, cost and complexity of computation remain substantial barriers to the use of NGS in routine clinical care. The clinical potential of NGS will not be realized until robust and routine whole genome sequencing data can be accurately rendered to medically actionable reports within a time window of hours and at scales of economy in the 10's of dollars.
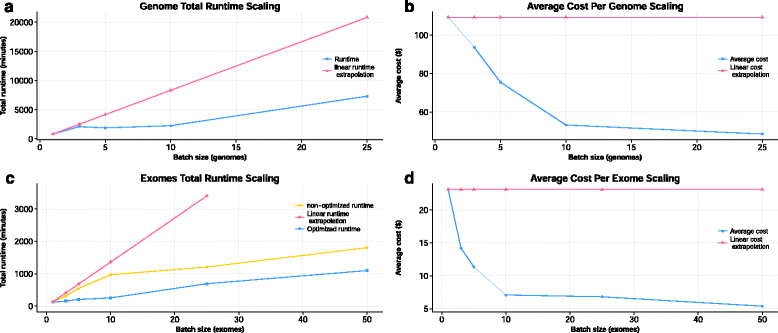
Results: We take a step towards addressing this challenge, by using COSMOS, a cloud-enabled workflow management system, to develop GenomeKey, an NGS whole genome analysis workflow. COSMOS implements complex workflows making optimal use of high-performance compute clusters. Here we show that the Amazon Web Service (AWS) implementation of GenomeKey via COSMOS provides a fast, scalable, and cost-effective analysis of both public benchmarking and large-scale heterogeneous clinical NGS datasets.
Conclusions: Our systematic benchmarking reveals important new insights and considerations to produce clinical turn-around of whole genome analysis optimization and workflow management including strategic batching of individual genomes and efficient cluster resource configuration.
Figures


References
-
- Life Technologies Receives FDA 510(k) Clearance for Diagnostic Use of Sanger Sequencing Platform and HLA Typing Kits [https://www.genomeweb.com/sequencing/510k-clearance-3500-dx-life-tech-ai...]
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

