Loss of DNA methylation at imprinted loci is a frequent event in hepatocellular carcinoma and identifies patients with shortened survival
- PMID: 26473022
- PMCID: PMC4606497
- DOI: 10.1186/s13148-015-0145-6
Loss of DNA methylation at imprinted loci is a frequent event in hepatocellular carcinoma and identifies patients with shortened survival
Abstract
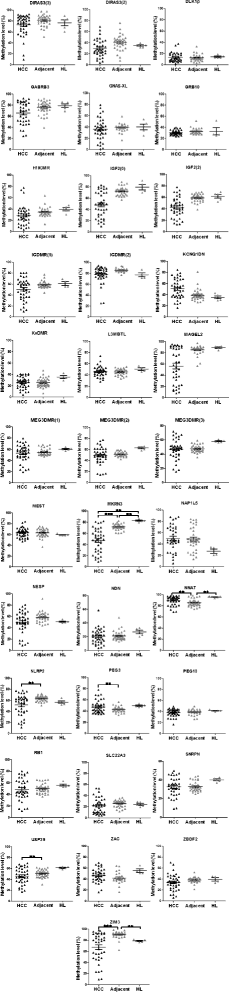
Background: Aberrant DNA methylation at imprinted loci is an important molecular mechanism contributing to several developmental and pathological disorders including cancer. However, knowledge about imprinting defects due to DNA methylation changes is relatively limited in hepatocellular carcinoma (HCC), the third leading cause of cancer death worldwide. Therefore, comprehensive quantitative DNA methylation analysis at imprinted loci showing ~50 % methylation in healthy liver tissues was performed in primary HCC specimens and the peritumoural liver tissues.
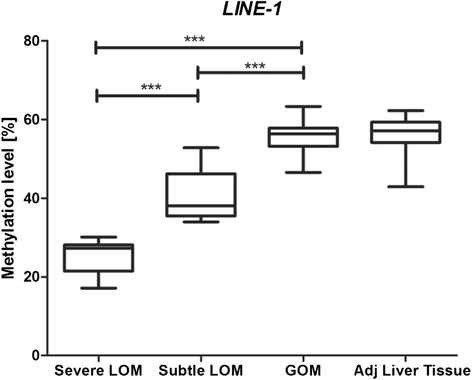
Results: We found frequent and extensive DNA methylation aberrations at many imprinted loci in HCC. Unsupervised cluster analysis of DNA methylation patterns at imprinted loci revealed subgroups of HCCs with moderate and severe loss of methylation. Hypomethylation at imprinted loci correlated significantly with poor overall survival (log-rank test, p = 0.02). Demethylation at imprinted loci was accompanied by loss of methylation at LINE-1, a commonly used marker for global DNA methylation levels (p < 0.001). In addition, we found that loss of methylation at imprinted loci correlated with the presence of a CTNNB1 mutation (Fisher's exact test p = 0.03). Re-analysis of publically available genome-wide methylation data sets confirmed our findings. The analysis of benign liver tumours (hepatocellular adenoma (HCA) and focal nodular hyperplasia (FNH)), the corresponding adjacent liver tissues, and healthy liver tissues showed that aberrant DNA methylation at imprinted loci is specific for HCC.
Conclusions: Our analyses demonstrate frequent and widespread DNA methylation aberrations at imprinted loci in human HCC and identified a hypomethylated subgroup of patients with shorter overall survival.
Keywords: DNA methylation; Hepatocellular carcinoma; Imprinting; Pyrosequencing.
Figures


Similar articles
-
LINE-1 hypomethylation in human hepatocellular carcinomas correlates with shorter overall survival and CIMP phenotype.PLoS One. 2019 May 6;14(5):e0216374. doi: 10.1371/journal.pone.0216374. eCollection 2019. PLoS One. 2019. PMID: 31059558 Free PMC article.
-
Deregulation of RB1 expression by loss of imprinting in human hepatocellular carcinoma.J Pathol. 2014 Aug;233(4):392-401. doi: 10.1002/path.4376. Epub 2014 Jun 16. J Pathol. 2014. PMID: 24838394
-
Loss of imprinting and allelic switching at the DLK1-MEG3 locus in human hepatocellular carcinoma.PLoS One. 2012;7(11):e49462. doi: 10.1371/journal.pone.0049462. Epub 2012 Nov 8. PLoS One. 2012. PMID: 23145177 Free PMC article.
-
Integrative analysis of aberrant Wnt signaling in hepatitis B virus-related hepatocellular carcinoma.World J Gastroenterol. 2015 May 28;21(20):6317-28. doi: 10.3748/wjg.v21.i20.6317. World J Gastroenterol. 2015. PMID: 26034368 Free PMC article. Review.
-
Disturbed methylation at multiple imprinted loci: an increasing observation in imprinting disorders.Epigenomics. 2011 Oct;3(5):625-37. doi: 10.2217/epi.11.84. Epigenomics. 2011. PMID: 22126250 Review.
Cited by
-
Transposable Elements in Human Cancer: Causes and Consequences of Deregulation.Int J Mol Sci. 2017 May 4;18(5):974. doi: 10.3390/ijms18050974. Int J Mol Sci. 2017. PMID: 28471386 Free PMC article. Review.
-
Human liver epigenetic alterations in non-alcoholic steatohepatitis are related to insulin action.Epigenetics. 2017 Apr 3;12(4):287-295. doi: 10.1080/15592294.2017.1294305. Epub 2017 Feb 23. Epigenetics. 2017. PMID: 28277977 Free PMC article.
-
Global Level of Plasma DNA Methylation is Associated with Overall Survival in Patients with Hepatocellular Carcinoma.Ann Surg Oncol. 2017 Nov;24(12):3788-3795. doi: 10.1245/s10434-017-5913-4. Epub 2017 Jun 7. Ann Surg Oncol. 2017. PMID: 28593503 Free PMC article.
-
LINE-1 hypomethylation in human hepatocellular carcinomas correlates with shorter overall survival and CIMP phenotype.PLoS One. 2019 May 6;14(5):e0216374. doi: 10.1371/journal.pone.0216374. eCollection 2019. PLoS One. 2019. PMID: 31059558 Free PMC article.
-
Downregulation of MGMT promotes proliferation of intrahepatic cholangiocarcinoma by regulating p21.Clin Transl Oncol. 2020 Mar;22(3):392-400. doi: 10.1007/s12094-019-02140-9. Epub 2019 Jul 1. Clin Transl Oncol. 2020. PMID: 31264147
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

