Differences in the Early Development of Human and Mouse Embryonic Stem Cells
- PMID: 26473594
- PMCID: PMC4608779
- DOI: 10.1371/journal.pone.0140803
Differences in the Early Development of Human and Mouse Embryonic Stem Cells
Abstract
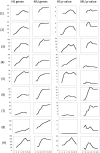
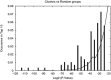
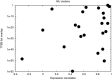
We performed a systematic analysis of gene expression features in early (10-21 days) development of human vs mouse embryonic cells (hESCs vs mESCs). Many development features were found to be conserved, and a majority of differentially regulated genes have similar expression change in both organisms. The similarity is especially evident, when gene expression profiles are clustered together and properties of clustered groups of genes are compared. First 10 days of mESC development match the features of hESC development within 21 days, in accordance with the differences in population doubling time in human and mouse ESCs. At the same time, several important differences are seen. There is a clear difference in initial expression change of transcription factors and stimulus responsive genes, which may be caused by the difference in experimental procedures. However, we also found that some biological processes develop differently; this can clearly be shown, for example, for neuron and sensory organ development. Some groups of genes show peaks of the expression levels during the development and these peaks cannot be claimed to happen at the same time points in the two organisms, as well as for the same groups of (orthologous) genes. We also detected a larger number of upregulated genes during development of mESCs as compared to hESCs. The differences were quantified by comparing promoters of related genes. Most of gene groups behave similarly and have similar transcription factor (TF) binding sites on their promoters. A few groups of genes have similar promoters, but are expressed differently in two species. Interestingly, there are groups of genes expressed similarly, although they have different promoters, which can be shown by comparing their TF binding sites. Namely, a large group of similarly expressed cell cycle-related genes is found to have discrepant TF binding properties in mouse vs human.
Conflict of interest statement
Figures




References
-
- Sato N, Sanjuan IM, Heke M, Uchida M, Naef F, Brivanlou AH. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev Biol. 2003;260(2):404–13. Epub 2003/08/19. doi: S0012160603002562 [pii]. . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

