Multi-Modal Imaging with a Toolbox of Influenza A Reporter Viruses
- PMID: 26473913
- PMCID: PMC4632381
- DOI: 10.3390/v7102873
Multi-Modal Imaging with a Toolbox of Influenza A Reporter Viruses
Abstract
Reporter viruses are useful probes for studying multiple stages of the viral life cycle. Here we describe an expanded toolbox of fluorescent and bioluminescent influenza A reporter viruses. The enhanced utility of these tools enabled kinetic studies of viral attachment, infection, and co-infection. Multi-modal bioluminescence and positron emission tomography-computed tomography (PET/CT) imaging of infected animals revealed that antiviral treatment reduced viral load, dissemination, and inflammation. These new technologies and applications will dramatically accelerate in vitro and in vivo influenza virus studies.
Keywords: influenza virus; multi-modal imaging; multiplicity reactivation; reporter virus.
Figures
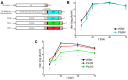
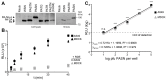
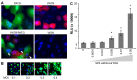

References
-
- Martinez-Sobrido L., Cadagan R., Steel J., Basler C.F., Palese P., Moran T.M., Garcia-Sastre A. Hemagglutinin-pseudotyped green fluorescent protein-expressing influenza viruses for the detection of influenza virus neutralizing antibodies. J. Virol. 2010;84:2157–2163. doi: 10.1128/JVI.01433-09. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

