The Candida albicans Histone Acetyltransferase Hat1 Regulates Stress Resistance and Virulence via Distinct Chromatin Assembly Pathways
- PMID: 26473952
- PMCID: PMC4608838
- DOI: 10.1371/journal.ppat.1005218
The Candida albicans Histone Acetyltransferase Hat1 Regulates Stress Resistance and Virulence via Distinct Chromatin Assembly Pathways
Abstract
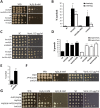
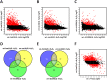
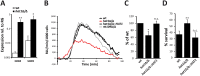
Human fungal pathogens like Candida albicans respond to host immune surveillance by rapidly adapting their transcriptional programs. Chromatin assembly factors are involved in the regulation of stress genes by modulating the histone density at these loci. Here, we report a novel role for the chromatin assembly-associated histone acetyltransferase complex NuB4 in regulating oxidative stress resistance, antifungal drug tolerance and virulence in C. albicans. Strikingly, depletion of the NuB4 catalytic subunit, the histone acetyltransferase Hat1, markedly increases resistance to oxidative stress and tolerance to azole antifungals. Hydrogen peroxide resistance in cells lacking Hat1 results from higher induction rates of oxidative stress gene expression, accompanied by reduced histone density as well as subsequent increased RNA polymerase recruitment. Furthermore, hat1Δ/Δ cells, despite showing growth defects in vitro, display reduced susceptibility to reactive oxygen-mediated killing by innate immune cells. Thus, clearance from infected mice is delayed although cells lacking Hat1 are severely compromised in killing the host. Interestingly, increased oxidative stress resistance and azole tolerance are phenocopied by the loss of histone chaperone complexes CAF-1 and HIR, respectively, suggesting a central role for NuB4 in the delivery of histones destined for chromatin assembly via distinct pathways. Remarkably, the oxidative stress phenotype of hat1Δ/Δ cells is a species-specific trait only found in C. albicans and members of the CTG clade. The reduced azole susceptibility appears to be conserved in a wider range of fungi. Thus, our work demonstrates how highly conserved chromatin assembly pathways can acquire new functions in pathogenic fungi during coevolution with the host.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
The histone acetyltransferase Hat1 facilitates DNA damage repair and morphogenesis in Candida albicans.Mol Microbiol. 2012 Dec;86(5):1197-214. doi: 10.1111/mmi.12051. Epub 2012 Oct 17. Mol Microbiol. 2012. PMID: 23075292
-
Epigenetic Control of Oxidative Stresses by Histone Acetyltransferases in Candida albicans.J Microbiol Biotechnol. 2018 Feb 28;28(2):181-189. doi: 10.4014/jmb.1707.07029. J Microbiol Biotechnol. 2018. PMID: 29169224 Review.
-
The Fungal Histone Acetyl Transferase Gcn5 Controls Virulence of the Human Pathogen Candida albicans through Multiple Pathways.Sci Rep. 2019 Jul 1;9(1):9445. doi: 10.1038/s41598-019-45817-5. Sci Rep. 2019. PMID: 31263212 Free PMC article.
-
Nucleosome assembly factors CAF-1 and HIR modulate epigenetic switching frequencies in an H3K56 acetylation-associated manner in Candida albicans.Eukaryot Cell. 2013 Apr;12(4):591-603. doi: 10.1128/EC.00334-12. Epub 2013 Feb 15. Eukaryot Cell. 2013. PMID: 23417560 Free PMC article.
-
Histone acetylation/deacetylation in Candida albicans and their potential as antifungal targets.Future Microbiol. 2020 Jul;15:1075-1090. doi: 10.2217/fmb-2019-0343. Epub 2020 Aug 28. Future Microbiol. 2020. PMID: 32854542 Review.
Cited by
-
Transcriptomics and Phenotyping Define Genetic Signatures Associated with Echinocandin Resistance in Candida auris.mBio. 2022 Aug 30;13(4):e0079922. doi: 10.1128/mbio.00799-22. Epub 2022 Aug 15. mBio. 2022. PMID: 35968956 Free PMC article.
-
Whole-genome sequencing of the invasive golden apple snail Pomacea canaliculata from Asia reveals rapid expansion and adaptive evolution.Gigascience. 2024 Jan 2;13:giae064. doi: 10.1093/gigascience/giae064. Gigascience. 2024. PMID: 39311763 Free PMC article.
-
Epigenetic mechanisms of drug resistance in fungi.Fungal Genet Biol. 2019 Nov;132:103253. doi: 10.1016/j.fgb.2019.103253. Epub 2019 Jul 17. Fungal Genet Biol. 2019. PMID: 31325489 Free PMC article. Review.
-
A J Domain Protein Functions as a Histone Chaperone to Maintain Genome Integrity and the Response to DNA Damage in a Human Fungal Pathogen.mBio. 2021 Dec 21;12(6):e0327321. doi: 10.1128/mbio.03273-21. Epub 2021 Dec 21. mBio. 2021. PMID: 34933457 Free PMC article.
-
Phototrophy and starvation-based induction of autophagy upon removal of Gcn5-catalyzed acetylation of Atg7 in Magnaporthe oryzae.Autophagy. 2017 Aug 3;13(8):1318-1330. doi: 10.1080/15548627.2017.1327103. Epub 2017 Jun 8. Autophagy. 2017. PMID: 28594263 Free PMC article.
References
-
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260. - PubMed
-
- Kaplan CD, Laprade L, Winston F (2003) Transcription elongation factors repress transcription initiation from cryptic sites. Science 301: 1096–1099. - PubMed
-
- Saunders A, Werner J, Andrulis ED, Nakayama T, Hirose S, et al. (2003) Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo . Science 301: 1094–1096. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

