New encouraging developments in contact prediction: Assessment of the CASP11 results
- PMID: 26474083
- PMCID: PMC4834069
- DOI: 10.1002/prot.24943
New encouraging developments in contact prediction: Assessment of the CASP11 results
Abstract
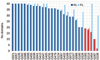
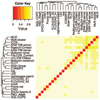
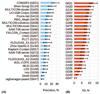
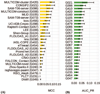
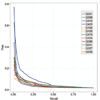
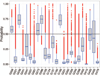
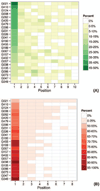
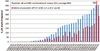
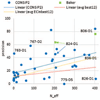
This article provides a report on the state-of-the-art in the prediction of intra-molecular residue-residue contacts in proteins based on the assessment of the predictions submitted to the CASP11 experiment. The assessment emphasis is placed on the accuracy in predicting long-range contacts. Twenty-nine groups participated in contact prediction in CASP11. At least eight of them used the recently developed evolutionary coupling techniques, with the top group (CONSIP2) reaching precision of 27% on target proteins that could not be modeled by homology. This result indicates a breakthrough in the development of methods based on the correlated mutation approach. Successful prediction of contacts was shown to be practically helpful in modeling three-dimensional structures; in particular target T0806 was modeled exceedingly well with accuracy not yet seen for ab initio targets of this size (>250 residues). Proteins 2016; 84(Suppl 1):131-144. © 2015 Wiley Periodicals, Inc.
Keywords: CASP; co-variation; contact prediction; correlated mutations; evolutionary coupling.
© 2015 Wiley Periodicals, Inc.
Figures

References
-
- Lesk AM. CASP2: report on ab initio predictions. Proteins. 1997;(Suppl 1):151–166. - PubMed
-
- Orengo CA, Bray JE, Hubbard T, LoConte L, Sillitoe I. Analysis and assessment of ab initio three-dimensional prediction, secondary structure, and contacts prediction. Proteins. 1999;(Suppl 3):149–170. - PubMed
-
- Lesk AM, Lo Conte L, Hubbard TJ. Assessment of novel fold targets in CASP4: predictions of three-dimensional structures, secondary structures, and interresidue contacts. Proteins. 2001;(Suppl 5):98–118. - PubMed
-
- Aloy P, Stark A, Hadley C, Russell RB. Predictions without templates: new folds, secondary structure, and contacts in CASP5. Proteins. 2003;53(Suppl 6):436–456. - PubMed
-
- Grana O, Baker D, MacCallum RM, Meiler J, Punta M, Rost B, Tress ML, Valencia A. CASP6 assessment of contact prediction. Proteins. 2005;61(Suppl 7):214–224. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

