Chronic hepatitis C viral infection subverts vaccine-induced T-cell immunity in humans
- PMID: 26474390
- PMCID: PMC4842008
- DOI: 10.1002/hep.28294
Chronic hepatitis C viral infection subverts vaccine-induced T-cell immunity in humans
Abstract
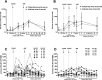
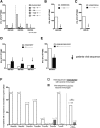
Adenoviral vectors encoding hepatitis C virus (HCV) nonstructural (NS) proteins induce multispecific, high-magnitude, durable CD4(+) and CD8(+) T-cell responses in healthy volunteers. We assessed the capacity of these vaccines to induce functional HCV-specific immune responses and determine T-cell cross-reactivity to endogenous virus in patients with chronic HCV infection. HCV genotype 1-infected patients were vaccinated using heterologous adenoviral vectors (ChAd3-NSmut and Ad6-NSmut) encoding HCV NS proteins in a dose escalation, prime-boost regimen, with and without concomitant pegylated interferon-α/ribavirin therapy. Analysis of immune responses ex vivo used human leukocyte antigen class I pentamers, intracellular cytokine staining, and fine mapping in interferon-γ enzyme-linked immunospot assays. Cross-reactivity of T cells with population and endogenous viral variants was determined following viral sequence analysis. Compared to healthy volunteers, the magnitude of HCV-specific T-cell responses following vaccination was markedly reduced. CD8(+) HCV-specific T-cell responses were detected in 15/24 patients at the highest dose, whereas CD4(+) T-cell responses were rarely detectable. Analysis of the host circulating viral sequence showed that T-cell responses were rarely elicited when there was sequence homology between vaccine immunogen and endogenous virus. In contrast, T cells were induced in the context of genetic mismatch between vaccine immunogen and endogenous virus; however, these commonly failed to recognize circulating epitope variants and had a distinct partially functional phenotype. Vaccination was well tolerated but had no significant effect on HCV viral load.
Conclusion: Vaccination with potent HCV adenoviral vectored vaccines fails to restore T-cell immunity except where there is genetic mismatch between vaccine immunogen and endogenous virus; this highlights the major challenge of overcoming T-cell exhaustion in the context of persistent antigen exposure with implications for cancer and other persistent infections.
© 2015 The Authors. HEPATOLOGY published by Wiley Periodicals, Inc., on behalf of the American Association for the Study of Liver Diseases.
Figures



Comment in
-
Vaccine-induced hepatitis C virus-specific CD8+ T cells do not always help.Hepatology. 2016 May;63(5):1411-4. doi: 10.1002/hep.28388. Epub 2016 Feb 19. Hepatology. 2016. PMID: 26662626 No abstract available.
References
-
- McKiernan SM, Hagan R, Curry M, McDonald GS, Kelly A, Nolan N, et al. Distinct MHC class I and II alleles are associated with hepatitis C viral clearance, originating from a single source. Hepatology 2004;40:108‐114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

