Caveolin-1-negative head and neck squamous cell carcinoma primary tumors display increased epithelial to mesenchymal transition and prometastatic properties
- PMID: 26474461
- PMCID: PMC4747196
- DOI: 10.18632/oncotarget.6099
Caveolin-1-negative head and neck squamous cell carcinoma primary tumors display increased epithelial to mesenchymal transition and prometastatic properties
Abstract
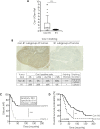
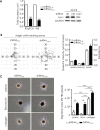
Distant metastases arise in 20-30% of patients with squamous cell carcinoma of the head and neck (HNSCC) in the 2 years following treatment. Therapeutic options are limited and the outcome of the patients is poor. The identification of predictive biomarkers of patient at risk for distant metastasis and therapies are urgently needed. We previously identified a clinical subgroup, called "R1" characterized by high propensity for rapid distant metastasis. Here, we showed that "R1" patients do not or at very low level express caveolin-1 (Cav1). Low or no expression of Cav1 is of bad prognosis. Disappearance of Cav1 enables cells to undergo epithelial-mesenchymal transition (EMT). EMT is associated with enhanced migration and invasion. Our study uncovered a new target, α5β1 integrin. Targeting α5β1 integrins might not only prevent metastasis of HNSCC but also delay the development of the primary tumor by reducing tumor cell viability. Cav1 detection might be taken into consideration in the future in the clinic not only to identify patients at high risk of metastasis but also to select patient who might benefit from an anti-integrin therapy.
Keywords: caveolin-1; head and neck cancer; integrins; metastasis.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures







Similar articles
-
Enhanced cell migration and apoptosis resistance may underlie the association between high SERPINE1 expression and poor outcome in head and neck carcinoma patients.Oncotarget. 2015 Oct 6;6(30):29016-33. doi: 10.18632/oncotarget.5032. Oncotarget. 2015. PMID: 26359694 Free PMC article.
-
Molecular profiling of tumour budding implicates TGFβ-mediated epithelial-mesenchymal transition as a therapeutic target in oral squamous cell carcinoma.J Pathol. 2015 Aug;236(4):505-16. doi: 10.1002/path.4550. Epub 2015 Jun 1. J Pathol. 2015. PMID: 25925492
-
Expression of IL-1α correlates with distant metastasis in patients with head and neck squamous cell carcinoma.Oncotarget. 2015 Nov 10;6(35):37398-409. doi: 10.18632/oncotarget.6054. Oncotarget. 2015. PMID: 26460957 Free PMC article.
-
Macrophage migration inhibitory factor: a potential driver and biomarker for head and neck squamous cell carcinoma.Oncotarget. 2017 Feb 7;8(6):10650-10661. doi: 10.18632/oncotarget.12890. Oncotarget. 2017. PMID: 27788497 Free PMC article. Review.
-
Epithelial-to-mesenchymal transition and cancer stem(-like) cells in head and neck squamous cell carcinoma.Cancer Lett. 2013 Sep 10;338(1):47-56. doi: 10.1016/j.canlet.2012.06.013. Epub 2012 Jul 4. Cancer Lett. 2013. PMID: 22771535 Review.
Cited by
-
Immunocompromised and immunocompetent mouse models for head and neck squamous cell carcinoma.Onco Targets Ther. 2016 Jan 27;9:545-55. doi: 10.2147/OTT.S95633. eCollection 2016. Onco Targets Ther. 2016. PMID: 26869799 Free PMC article. Review.
-
Platelet Integrins in Tumor Metastasis: Do They Represent a Therapeutic Target?Cancers (Basel). 2017 Sep 28;9(10):133. doi: 10.3390/cancers9100133. Cancers (Basel). 2017. PMID: 28956830 Free PMC article. Review.
-
Differences in the expression of caveolin-1 isoforms in cancer-associated and normal fibroblasts of patients with oral squamous cell carcinoma.Clin Oral Investig. 2021 Oct;25(10):5823-5831. doi: 10.1007/s00784-021-03887-8. Epub 2021 Mar 27. Clin Oral Investig. 2021. PMID: 33774714 Free PMC article.
-
Involvement of the TGF-β1 pathway in caveolin-1-associated regulation of head and neck tumor cell metastasis.Oncol Lett. 2020 Feb;19(2):1298-1304. doi: 10.3892/ol.2019.11187. Epub 2019 Dec 6. Oncol Lett. 2020. PMID: 31966060 Free PMC article.
-
Cav1/EREG/YAP Axis in the Treatment Resistance of Cav1-Expressing Head and Neck Squamous Cell Carcinoma.Cancers (Basel). 2021 Jun 18;13(12):3038. doi: 10.3390/cancers13123038. Cancers (Basel). 2021. PMID: 34207120 Free PMC article.
References
-
- Hashibe M, Brennan P, Chuang SC, Boccia S, Castellsague X, Chen C, Curado MP, Dal Maso L, Daudt AW, Fabianova E, Fernandez L, Wunsch-Filho V, Franceschi S, Hayes RB, Herrero R, Kelsey K, et al. Interaction between tobacco and alcohol use and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2009;18:541–550. - PMC - PubMed
-
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61:69–90. - PubMed
-
- Denaro N, Russi EG, Adamo V, Merlano MC. State-of-the-art and emerging treatment options in the management of head and neck cancer: news from 2013. Oncology. 2014;86:212–229. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

