A Phenomic Scan of the Norfolk Island Genetic Isolate Identifies a Major Pleiotropic Effect Locus Associated with Metabolic and Renal Disorder Markers
- PMID: 26474483
- PMCID: PMC4608754
- DOI: 10.1371/journal.pgen.1005593
A Phenomic Scan of the Norfolk Island Genetic Isolate Identifies a Major Pleiotropic Effect Locus Associated with Metabolic and Renal Disorder Markers
Abstract
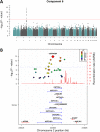
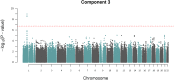
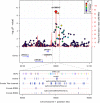
Multiphenotype genome-wide association studies (GWAS) may reveal pleiotropic genes, which would remain undetected using single phenotype analyses. Analysis of large pedigrees offers the added advantage of more accurately assessing trait heritability, which can help prioritise genetically influenced phenotypes for GWAS analysis. In this study we performed a principal component analysis (PCA), heritability (h2) estimation and pedigree-based GWAS of 37 cardiovascular disease -related phenotypes in 330 related individuals forming a large pedigree from the Norfolk Island genetic isolate. PCA revealed 13 components explaining >75% of the total variance. Nine components yielded statistically significant h2 values ranging from 0.22 to 0.54 (P<0.05). The most heritable component was loaded with 7 phenotypic measures reflecting metabolic and renal dysfunction. A GWAS of this composite phenotype revealed statistically significant associations for 3 adjacent SNPs on chromosome 1p22.2 (P<1x10-8). These SNPs form a 42kb haplotype block and explain 11% of the genetic variance for this renal function phenotype. Replication analysis of the tagging SNP (rs1396315) in an independent US cohort supports the association (P = 0.000011). Blood transcript analysis showed 35 genes were associated with rs1396315 (P<0.05). Gene set enrichment analysis of these genes revealed the most enriched pathway was purine metabolism (P = 0.0015). Overall, our findings provide convincing evidence for a major pleiotropic effect locus on chromosome 1p22.2 influencing risk of renal dysfunction via purine metabolism pathways in the Norfolk Island population. Further studies are now warranted to interrogate the functional relevance of this locus in terms of renal pathology and cardiovascular disease risk.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Laslett LJ, Alagona P, Clark BA, Drozda JP, Saldivar F, Wilson SR, et al. The worldwide environment of cardiovascular disease: prevalence, diagnosis, therapy, and policy issues: a report from the American College of Cardiology. J Am Coll Cardiol. 2012;60: S1–49. 10.1016/j.jacc.2012.11.002 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

