Expression Quantitative Trait Loci Information Improves Predictive Modeling of Disease Relevance of Non-Coding Genetic Variation
- PMID: 26474488
- PMCID: PMC4608673
- DOI: 10.1371/journal.pone.0140758
Expression Quantitative Trait Loci Information Improves Predictive Modeling of Disease Relevance of Non-Coding Genetic Variation
Abstract
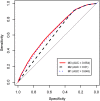
Disease-associated loci identified through genome-wide association studies (GWAS) frequently localize to non-coding sequence. We and others have demonstrated strong enrichment of such single nucleotide polymorphisms (SNPs) for expression quantitative trait loci (eQTLs), supporting an important role for regulatory genetic variation in complex disease pathogenesis. Herein we describe our initial efforts to develop a predictive model of disease-associated variants leveraging eQTL information. We first catalogued cis-acting eQTLs (SNPs within 100 kb of target gene transcripts) by meta-analyzing four studies of three blood-derived tissues (n = 586). At a false discovery rate < 5%, we mapped eQTLs for 6,535 genes; these were enriched for disease-associated genes (P < 10(-04)), particularly those related to immune diseases and metabolic traits. Based on eQTL information and other variant annotations (distance from target gene transcript, minor allele frequency, and chromatin state), we created multivariate logistic regression models to predict SNP membership in reported GWAS. The complete model revealed independent contributions of specific annotations as strong predictors, including evidence for an eQTL (odds ratio (OR) = 1.2-2.0, P < 10(-11)) and the chromatin states of active promoters, different classes of strong or weak enhancers, or transcriptionally active regions (OR = 1.5-2.3, P < 10(-11)). This complete prediction model including eQTL association information ultimately allowed for better discrimination of SNPs with higher probabilities of GWAS membership (6.3-10.0%, compared to 3.5% for a random SNP) than the other two models excluding eQTL information. This eQTL-based prediction model of disease relevance can help systematically prioritize non-coding GWAS SNPs for further functional characterization.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

