Evolution of Cis-Regulatory Elements and Regulatory Networks in Duplicated Genes of Arabidopsis
- PMID: 26474639
- PMCID: PMC4677880
- DOI: 10.1104/pp.15.00717
Evolution of Cis-Regulatory Elements and Regulatory Networks in Duplicated Genes of Arabidopsis
Abstract
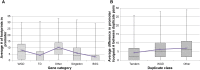
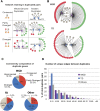
Plant genomes contain large numbers of duplicated genes that contribute to the evolution of new functions. Following duplication, genes can exhibit divergence in their coding sequence and their expression patterns. Changes in the cis-regulatory element landscape can result in changes in gene expression patterns. High-throughput methods developed recently can identify potential cis-regulatory elements on a genome-wide scale. Here, we use a recent comprehensive data set of DNase I sequencing-identified cis-regulatory binding sites (footprints) at single-base-pair resolution to compare binding sites and network connectivity in duplicated gene pairs in Arabidopsis (Arabidopsis thaliana). We found that duplicated gene pairs vary greatly in their cis-regulatory element architecture, resulting in changes in regulatory network connectivity. Whole-genome duplicates (WGDs) have approximately twice as many footprints in their promoters left by potential regulatory proteins than do tandem duplicates (TDs). The WGDs have a greater average number of footprint differences between paralogs than TDs. The footprints, in turn, result in more regulatory network connections between WGDs and other genes, forming denser, more complex regulatory networks than shown by TDs. When comparing regulatory connections between duplicates, WGDs had more pairs in which the two genes are either partially or fully diverged in their network connections, but fewer genes with no network connections than the TDs. There is evidence of younger TDs and WGDs having fewer unique connections compared with older duplicates. This study provides insights into cis-regulatory element evolution and network divergence in duplicated genes.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures

References
-
- Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 - PubMed
-
- Chung JH, Whiteley M, Felsenfeld G (1993) A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74: 505–514 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

