The development and application of a molecular community profiling strategy to identify polymicrobial bacterial DNA in the whole blood of septic patients
- PMID: 26474751
- PMCID: PMC4609058
- DOI: 10.1186/s12866-015-0557-7
The development and application of a molecular community profiling strategy to identify polymicrobial bacterial DNA in the whole blood of septic patients
Abstract
Background: The application of molecular based diagnostics in sepsis has had limited success to date. Molecular community profiling methods have indicated that polymicrobial infections are more common than suggested by standard clinical culture. A molecular profiling approach was developed to investigate the propensity for polymicrobial infections in patients predicted to have bacterial sepsis.
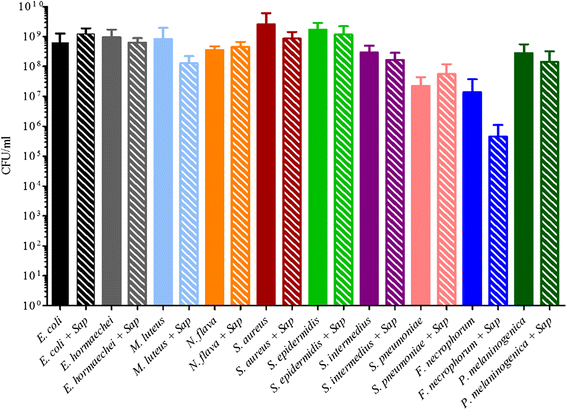
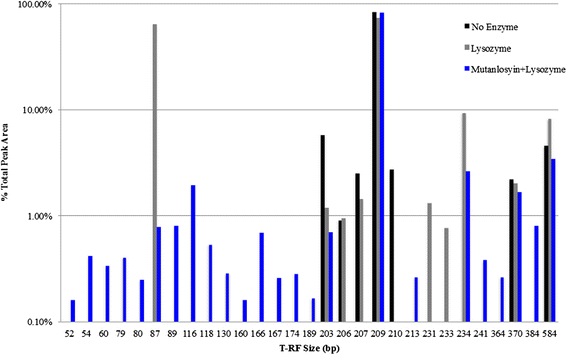
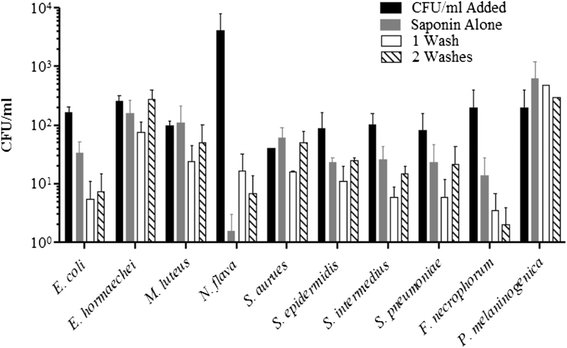
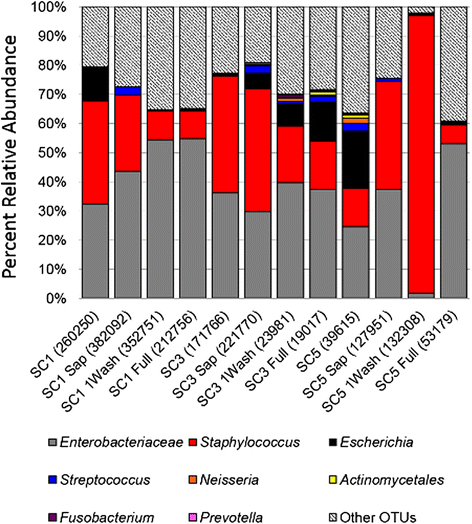
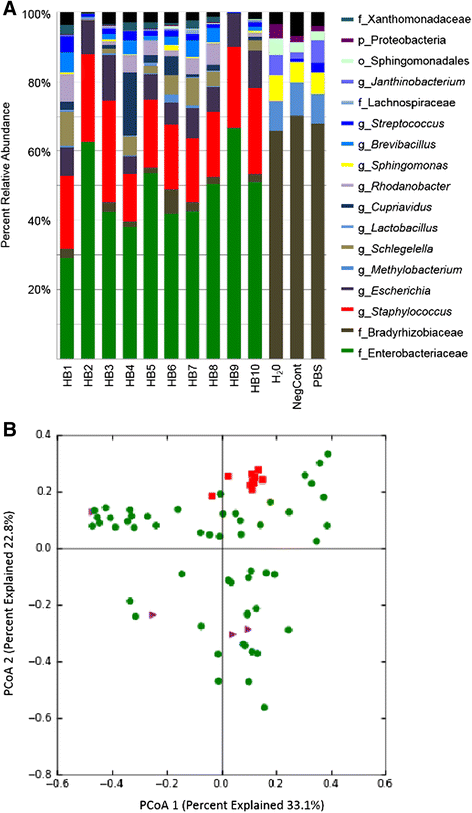
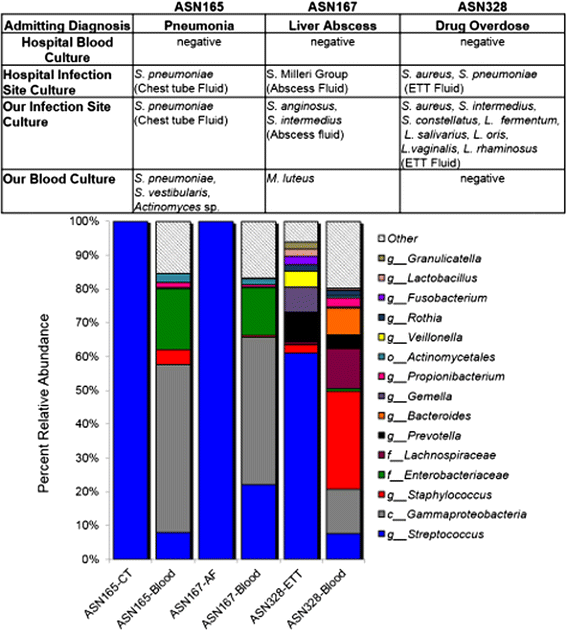
Results: Disruption of blood cells with saponin and hypotonic shock enabled the recovery of microbial cells with no significant changes in microbial growth when compared to CFU/ml values immediately prior to the addition of saponin. DNA extraction included a cell-wall digestion step with both lysozyme and mutanolysin, which increased the recovery of terminal restriction fragments by 2.4 fold from diverse organisms. Efficiencies of recovery and limits of detection using Illumina sequencing of the 16S rRNA V3 region were determined for both viable cells and DNA using mock bacterial communities inoculated into whole blood. Bacteria from pre-defined communities could be recovered following lysis and removal of host cells with >97% recovery of total DNA present. Applying the molecular profiling methodology to three septic patients in the intensive care unit revealed microbial DNA from blood had consistent alignment with cultured organisms from the primary infection site providing evidence for a bloodstream infection in the absence of a clinical lab positive blood culture result in two of the three cases. In addition, the molecular profiling indicated greater diversity was present in the primary infection sample when compared to clinical diagnostic culture.
Conclusions: A method for analyzing bacterial DNA from whole blood was developed in order to characterize the bacterial DNA profile of sepsis infections. Preliminary results indicated that sepsis infections were polymicrobial in nature with the bacterial DNA recovered suggesting a more complex etiology when compared to blood culture data.
Figures
Similar articles
-
Bacterial DNA patterns identified using paired-end Illumina sequencing of 16S rRNA genes from whole blood samples of septic patients in the emergency room and intensive care unit.BMC Microbiol. 2018 Jul 25;18(1):79. doi: 10.1186/s12866-018-1211-y. BMC Microbiol. 2018. PMID: 30045694 Free PMC article.
-
Evaluation of real-time PCR and pyrosequencing for screening incubating blood culture bottles from adults with suspected bloodstream infection.Diagn Microbiol Infect Dis. 2015 Mar;81(3):158-62. doi: 10.1016/j.diagmicrobio.2014.11.014. Epub 2014 Dec 3. Diagn Microbiol Infect Dis. 2015. PMID: 25534615 Free PMC article.
-
Molecular diagnosis of polymicrobial brain abscesses with 16S-rDNA-based next-generation sequencing.Clin Microbiol Infect. 2021 Jan;27(1):76-82. doi: 10.1016/j.cmi.2020.03.028. Epub 2020 Mar 31. Clin Microbiol Infect. 2021. PMID: 32244052
-
A 'culture' shift: Application of molecular techniques for diagnosing polymicrobial infections.Biotechnol Adv. 2019 May-Jun;37(3):476-490. doi: 10.1016/j.biotechadv.2019.02.013. Epub 2019 Feb 20. Biotechnol Adv. 2019. PMID: 30797092 Free PMC article. Review.
-
Microbial diagnosis of bloodstream infection: towards molecular diagnosis directly from blood.Clin Microbiol Infect. 2015 Apr;21(4):323-31. doi: 10.1016/j.cmi.2015.02.005. Epub 2015 Feb 14. Clin Microbiol Infect. 2015. PMID: 25686695 Review.
Cited by
-
Fast I(n)dentification of Pathogens in Neonates (FINDPATH-N): protocol for a prospective pilot cohort study of next-generation sequencing for pathogen identification in neonates with suspected sepsis.BMJ Paediatr Open. 2020 Apr 6;4(1):e000651. doi: 10.1136/bmjpo-2020-000651. eCollection 2020. BMJ Paediatr Open. 2020. PMID: 32518844 Free PMC article.
-
Diagnosis of Bacterial Bloodstream Infections: A 16S Metagenomics Approach.PLoS Negl Trop Dis. 2016 Feb 29;10(2):e0004470. doi: 10.1371/journal.pntd.0004470. eCollection 2016 Feb. PLoS Negl Trop Dis. 2016. PMID: 26927306 Free PMC article.
-
Bacterial DNA patterns identified using paired-end Illumina sequencing of 16S rRNA genes from whole blood samples of septic patients in the emergency room and intensive care unit.BMC Microbiol. 2018 Jul 25;18(1):79. doi: 10.1186/s12866-018-1211-y. BMC Microbiol. 2018. PMID: 30045694 Free PMC article.
-
Longitudinal variability in the urinary microbiota of healthy premenopausal women and the relation to neighboring microbial communities: A pilot study.PLoS One. 2022 Jan 14;17(1):e0262095. doi: 10.1371/journal.pone.0262095. eCollection 2022. PLoS One. 2022. PMID: 35030190 Free PMC article.
-
Pylephlebitis: A Systematic Review on Etiology, Diagnosis, and Treatment of Infective Portal Vein Thrombosis.Diagnostics (Basel). 2023 Jan 25;13(3):429. doi: 10.3390/diagnostics13030429. Diagnostics (Basel). 2023. PMID: 36766534 Free PMC article. Review.
References
-
- Valles J. [Bacteremias in intensive care] Enferm Infecc Microbiol Clin. 1997;15(Suppl 3):8–13. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

