GERV: a statistical method for generative evaluation of regulatory variants for transcription factor binding
- PMID: 26476779
- PMCID: PMC5860000
- DOI: 10.1093/bioinformatics/btv565
GERV: a statistical method for generative evaluation of regulatory variants for transcription factor binding
Abstract
Motivation: The majority of disease-associated variants identified in genome-wide association studies reside in noncoding regions of the genome with regulatory roles. Thus being able to interpret the functional consequence of a variant is essential for identifying causal variants in the analysis of genome-wide association studies.
Results: We present GERV (generative evaluation of regulatory variants), a novel computational method for predicting regulatory variants that affect transcription factor binding. GERV learns a k-mer-based generative model of transcription factor binding from ChIP-seq and DNase-seq data, and scores variants by computing the change of predicted ChIP-seq reads between the reference and alternate allele. The k-mers learned by GERV capture more sequence determinants of transcription factor binding than a motif-based approach alone, including both a transcription factor's canonical motif and associated co-factor motifs. We show that GERV outperforms existing methods in predicting single-nucleotide polymorphisms associated with allele-specific binding. GERV correctly predicts a validated causal variant among linked single-nucleotide polymorphisms and prioritizes the variants previously reported to modulate the binding of FOXA1 in breast cancer cell lines. Thus, GERV provides a powerful approach for functionally annotating and prioritizing causal variants for experimental follow-up analysis.
Availability and implementation: The implementation of GERV and related data are available at http://gerv.csail.mit.edu/.
© The Author 2015. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures

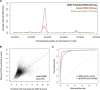

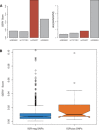
Similar articles
-
On the identification of potential regulatory variants within genome wide association candidate SNP sets.BMC Med Genomics. 2014 Jun 11;7:34. doi: 10.1186/1755-8794-7-34. BMC Med Genomics. 2014. PMID: 24920305 Free PMC article.
-
SeqGL Identifies Context-Dependent Binding Signals in Genome-Wide Regulatory Element Maps.PLoS Comput Biol. 2015 May 27;11(5):e1004271. doi: 10.1371/journal.pcbi.1004271. eCollection 2015 May. PLoS Comput Biol. 2015. PMID: 26016777 Free PMC article.
-
Which Genetics Variants in DNase-Seq Footprints Are More Likely to Alter Binding?PLoS Genet. 2016 Feb 22;12(2):e1005875. doi: 10.1371/journal.pgen.1005875. eCollection 2016 Feb. PLoS Genet. 2016. PMID: 26901046 Free PMC article.
-
Exploring the function of genetic variants in the non-coding genomic regions: approaches for identifying human regulatory variants affecting gene expression.Brief Bioinform. 2015 May;16(3):393-412. doi: 10.1093/bib/bbu018. Epub 2014 Jun 10. Brief Bioinform. 2015. PMID: 24916300 Review.
-
Mining the Unknown: Assigning Function to Noncoding Single Nucleotide Polymorphisms.Trends Genet. 2017 Jan;33(1):34-45. doi: 10.1016/j.tig.2016.10.008. Epub 2016 Dec 6. Trends Genet. 2017. PMID: 27939749 Free PMC article. Review.
Cited by
-
Identifying functional regulatory mutation blocks by integrating genome sequencing and transcriptome data.iScience. 2023 Jul 3;26(8):107266. doi: 10.1016/j.isci.2023.107266. eCollection 2023 Aug 18. iScience. 2023. PMID: 37520692 Free PMC article.
-
DNA Motif Recognition Modeling from Protein Sequences.iScience. 2018 Sep 28;7:198-211. doi: 10.1016/j.isci.2018.09.003. Epub 2018 Sep 10. iScience. 2018. PMID: 30267681 Free PMC article.
-
Recurrent Neural Network for Predicting Transcription Factor Binding Sites.Sci Rep. 2018 Oct 15;8(1):15270. doi: 10.1038/s41598-018-33321-1. Sci Rep. 2018. PMID: 30323198 Free PMC article.
-
GARLIC: a bioinformatic toolkit for aetiologically connecting diseases and cell type-specific regulatory maps.Hum Mol Genet. 2017 Feb 15;26(4):742-752. doi: 10.1093/hmg/ddw423. Hum Mol Genet. 2017. PMID: 28007912 Free PMC article.
-
Imputation for transcription factor binding predictions based on deep learning.PLoS Comput Biol. 2017 Feb 24;13(2):e1005403. doi: 10.1371/journal.pcbi.1005403. eCollection 2017 Feb. PLoS Comput Biol. 2017. PMID: 28234893 Free PMC article.
References
-
- Bartels M., et al. (2007) Peptide-mediated disruption of NFkappaB/NRF interaction inhibits IL-8 gene activation by IL-1 or Helicobacter pylori. J. Immunol., 179, 7605–7613. - PubMed
-
- Carroll J.S., et al. (2005) Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell, 122, 33–43. - PubMed
-
- Carroll J.S., et al. (2006) Genome-wide analysis of estrogen receptor binding sites. Nat. Genet., 38, 1289–1297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

