GERV: a statistical method for generative evaluation of regulatory variants for transcription factor binding
- PMID: 26476779
- PMCID: PMC5860000
- DOI: 10.1093/bioinformatics/btv565
GERV: a statistical method for generative evaluation of regulatory variants for transcription factor binding
Abstract
Motivation: The majority of disease-associated variants identified in genome-wide association studies reside in noncoding regions of the genome with regulatory roles. Thus being able to interpret the functional consequence of a variant is essential for identifying causal variants in the analysis of genome-wide association studies.
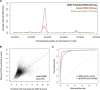
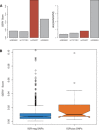
Results: We present GERV (generative evaluation of regulatory variants), a novel computational method for predicting regulatory variants that affect transcription factor binding. GERV learns a k-mer-based generative model of transcription factor binding from ChIP-seq and DNase-seq data, and scores variants by computing the change of predicted ChIP-seq reads between the reference and alternate allele. The k-mers learned by GERV capture more sequence determinants of transcription factor binding than a motif-based approach alone, including both a transcription factor's canonical motif and associated co-factor motifs. We show that GERV outperforms existing methods in predicting single-nucleotide polymorphisms associated with allele-specific binding. GERV correctly predicts a validated causal variant among linked single-nucleotide polymorphisms and prioritizes the variants previously reported to modulate the binding of FOXA1 in breast cancer cell lines. Thus, GERV provides a powerful approach for functionally annotating and prioritizing causal variants for experimental follow-up analysis.
Availability and implementation: The implementation of GERV and related data are available at http://gerv.csail.mit.edu/.
© The Author 2015. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures


References
-
- Bartels M., et al. (2007) Peptide-mediated disruption of NFkappaB/NRF interaction inhibits IL-8 gene activation by IL-1 or Helicobacter pylori. J. Immunol., 179, 7605–7613. - PubMed
-
- Carroll J.S., et al. (2005) Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell, 122, 33–43. - PubMed
-
- Carroll J.S., et al. (2006) Genome-wide analysis of estrogen receptor binding sites. Nat. Genet., 38, 1289–1297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

