Molecular logic of neuronal self-recognition through protocadherin domain interactions
- PMID: 26478182
- PMCID: PMC4624033
- DOI: 10.1016/j.cell.2015.09.026
Molecular logic of neuronal self-recognition through protocadherin domain interactions
Abstract
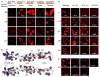
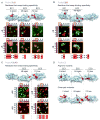
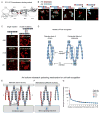
Self-avoidance, a process preventing interactions of axons and dendrites from the same neuron during development, is mediated in vertebrates through the stochastic single-neuron expression of clustered protocadherin protein isoforms. Extracellular cadherin (EC) domains mediate isoform-specific homophilic binding between cells, conferring cell recognition through a poorly understood mechanism. Here, we report crystal structures for the EC1-EC3 domain regions from four protocadherin isoforms representing the α, β, and γ subfamilies. All are rod shaped and monomeric in solution. Biophysical measurements, cell aggregation assays, and computational docking reveal that trans binding between cells depends on the EC1-EC4 domains, which interact in an antiparallel orientation. We also show that the EC6 domains are required for the formation of cis-dimers. Overall, our results are consistent with a model in which protocadherin cis-dimers engage in a head-to-tail interaction between EC1-EC4 domains from apposed cell surfaces, possibly forming a zipper-like protein assembly, and thus providing a size-dependent self-recognition mechanism.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures



References
-
- Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

