Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly
- PMID: 26479319
- PMCID: PMC4663675
- DOI: 10.1038/ncb3257
Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly
Abstract
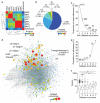
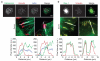
Integrin receptor activation initiates the formation of integrin adhesion complexes (IACs) at the cell membrane that transduce adhesion-dependent signals to control a multitude of cellular functions. Proteomic analyses of isolated IACs have revealed an unanticipated molecular complexity; however, a global view of the consensus composition and dynamics of IACs is lacking. Here, we have integrated several IAC proteomes and generated a 2,412-protein integrin adhesome. Analysis of this data set reveals the functional diversity of proteins in IACs and establishes a consensus adhesome of 60 proteins. The consensus adhesome is likely to represent a core cell adhesion machinery, centred around four axes comprising ILK-PINCH-kindlin, FAK-paxillin, talin-vinculin and α-actinin-zyxin-VASP, and includes underappreciated IAC components such as Rsu-1 and caldesmon. Proteomic quantification of IAC assembly and disassembly detailed the compositional dynamics of the core cell adhesion machinery. The definition of this consensus view of integrin adhesome components provides a resource for the research community.
Figures


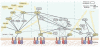



References
-
- Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. - PubMed
-
- Liu S, Calderwood DA, Ginsberg MH. Integrin cytoplasmic domain-binding proteins. J. Cell Sci. 2000;113:3563–3571. - PubMed
-
- Winograd-Katz SE, Fässler R, Geiger B, Legate KR. The integrin adhesome: from genes and proteins to human disease. Nat. Rev. Mol. Cell Biol. 2014;15:273–288. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

